Abstract
Axonal injury increases intracellular Ca2+ and cAMP and has been shown to induce gene expression, which is thought to be a key event for regeneration. Increases in intracellular Ca2+ and/or cAMP can alter gene expression via activation of a family of transcription factors that bind to and modulate the expression of CRE (Ca2+/cAMP response element) sequence-containing genes. We have used Aplysia motor neurons to examine the role of CRE-binding proteins in axonal regeneration after injury. We report that axonal injury increases the binding of proteins to a CRE sequence-containing probe. In addition, Western blot analysis revealed that the level of ApCREB2, a CRE sequence-binding repressor, was enhanced as a result of axonal injury. The sequestration of CRE-binding proteins by microinjection of CRE sequence-containing plasmids enhanced axon collateral formation (both number and length) as compared with control plasmid injections. These findings show that Ca2+/cAMP-mediated gene expression via CRE-binding transcription factors participates in the regeneration of motor neuron axons.
Regeneration of motor neuron axons in the central nervous system (CNS) generally is abortive and fails to reestablish functional connections (1). Both intrinsic genetic programs as well as environmental factors are thought to regulate regeneration in the CNS (2). After axonal injury, the distal segment undergoes a progressive demise called wallerian or anterograde degeneration. The proximal axon initially retracts followed by the emergence of axon collaterals. Collateral elaboration is thought to be a key event in the navigation of axons to the proper target during axonal regeneration as well as during the development of the CNS (3, 4). This process appears to involve alterations in gene expression and protein synthesis (5–11). However, the genomic programs that mediate collateral formation are yet to be identified. An understanding of this process would be of immense help in the development of therapies to facilitate motor neuron regeneration in humans.
Calcium is an important second messenger that plays a role in a variety of cellular functions. Axonal injury increases intracellular calcium concentrations to high levels (12). This increase in intracellular calcium is thought to result from calcium influx through both the exposed axon end and calcium channels. Increased levels of calcium can stimulate various enzymes including calcium-sensitive adenylyl cyclase [leading to cAMP-dependent protein kinase A (PKA) activation via increased intracellular cAMP], calcium/calmodulin-dependent protein kinases (CaMK), protein kinase C (PKC), mitogen-activated protein kinase (MAPK) and the protein phosphatase calcineurin (13–17). Stimulation of these calcium-sensitive enzymes can activate transcription factors via phosphorylation or dephosphorylation (18). These activated transcription factors, in turn, alter the expression of effector genes. Recent experiments have implicated Ca2+/cAMP-mediated gene expression, specifically the balance between CRE (Ca2+/cAMP response element)-binding activator and repressor functions, as a key event underlying morphological changes in neurons associated with long-term memory storage (19–26).
Most of the genes that are induced by Ca2+/cAMP contain one or more CRE (e.g., TGACGTCA) sequences in their promoter regions (27, 28). A family of transcription factors have been identified that can bind to the mammalian CRE sequence (29, 30, 49). These factors can form homo- or heterodimers with different degrees of activating or repressing function. Examples of activators are CREB (CRE-binding protein), CREM τ (CRE modulator τ), and ATF-1 (activating transcription factor-1), whereas repressors include ICER (inducible cAMP early repressor) and CREM α, β, and γ. The activator CREB has been shown to be phosphorylated by PKA, CaMK, or p90rsk (90-kDa protein ribosomal S6 kinase, also called CREB kinase), which enhances its transcriptional activity (17, 29, 31–34). In contrast, repressors lack the transactivation domain allowing them to bind to the CRE sequence without stimulating the transcriptional machinery (30).
Recently in Aplysia californica, a homolog of the activator CREB (ApCREB1) and a repressor (ApCREB2) have been cloned and characterized (21, 35). These factors are constitutively present and have been shown to be phosphorylated in response to agents that increase intracellular cAMP levels. However, other Aplysia CREB family members have not yet been isolated. The large and identifiable neurons of Aplysia allow manipulations within single neurons to investigate the molecular basis of cellular and morphological changes. In this study, we have taken advantage of these properties to examine the role of CRE-binding proteins in the regeneration of Aplysia tail motor neuron axons. To accomplish this, we performed microinjection studies by using CRE-binding sequence-containing plasmids as transcription factor decoys (TFDs) to sequester CRE-binding proteins.
EXPERIMENTAL PROCEDURES
Construction of Plasmid DNA.
Five copies of an oligonucleotide containing the CRE sequence (5′-GGCCTCCTTGGCTGACGTCAGAGAGAGAGTTCTGCA-3′) from the rat somatostatin gene or five copies of an oligonucleotide containing the TRE sequence (5′-GATCCAAGCAATTATGAGTCAGTTTGCCTGCA-3′) from the rat stromelysin gene were cloned into Bluescript plasmids (Stratagene) and purified by using standard techniques (36). The DNA was then dissolved in diethylpyrocarbonate (DEPC)-treated water and stored at 4°C.
Preparation of Pedal Ganglia Culture.
Aplysia californica (100–250 g) were obtained from Alacrity Marine Biological (Redondo Beach, CA). Animals were maintained in artificial sea water (Instant Ocean) at 15°C for at least 5 days before any experimental manipulation. Animals were anesthetized by injecting one-third their body weight of cold isotonic MgCl2 (363 mM) into the neck region and kept in artificial sea water maintained at 0°C for 10 min. The ring ganglion (cerebral ganglion, right and left pleural and pedal ganglia) was dissected out with the connectives and nerves being cut far away from the ganglia. The pedal ganglia were desheathed while submerged in L15 (Leibowitz 15, Sigma) containing 208.5 mM MgCl2 to expose the tail motor neurons. One randomly chosen pedal ganglion was used as the experimental ganglion and its symmetrical partner was used as control. The ganglia were rinsed several times with L15 and rested for 2–3 hr before microinjection.
Microinjection of Plasmid DNA.
Plasmid DNA solutions were prepared at a concentration of 100 μg/ml in 0.5 M potassium acetate and 0.05% Fast green. The resulting solution was centrifuged and used to fill a glass electrode that was beveled to a resistance of 20 MΩ. A tail motor neuron, identified by its position on the pedal ganglion, was impaled with a glass electrode filled with either plasmids or saline. To confirm that the impaled motor neuron axon is projecting through the p9 nerve, p9 was stimulated to generate antidromic spikes. A Picospritzer (General Valve, Fairfield, NJ) was used for microinjection using 5 × 5-msec pressure pulses. The volume of the injected liquid was estimated to be 50 pl. This corresponds to approximately 1 × 106 copies of plasmid DNA injected per neuron. Because Aplysia neurons are highly polypoidal (37, 38), a large copy number of TFDs was used for these experiments.
Axonal Crush.
Approximately 15 min after the microinjection of plasmid DNA, the p9 pedal nerves for both the experimental and its paired control were crushed under a dissection microscope by using fine forceps (10). The crush transected the axons but not the surrounding sheath. Although the removal of ganglia from the animal required transection of all nerves, the crush was used to provide a standardized injury paradigm. Moreover, this allowed visualization of the regenerating axons within a defined area. After the crush, ganglia were placed in a medium composed of 1:1 L15 and Aplysia hemolymph. Ganglia were maintained in a 16°C incubator for 1 week before visualization.
Morphological Visualization of Axons.
The injected motor neurons were identified by the presence of Fast green. The viability of these neurons was assessed by measuring the input resistance and generating action potentials. Healthy motor neurons were injected with rhodamine-dextran to visualize their axons. The dye was allowed to fill the axons overnight. The ganglia then were fixed overnight by using 4% paraformaldehyde in PBS and visualized by using epifluorescent illumination (Zeiss Axiophot) with a rhodamine filter set. The length and number of axon collaterals were determined by the use of neurolucida (MicroBrightField, Colchester, VT) by an independent observer (39). The neurolucida program works by superimposing a computer screen over the microscopic image. This allows the observer to trace each axon collateral by marking points along its length and sizing the mouse-controlled cursor to represent the width of each segment. A reference point allows the traced image to move simultaneously with the microscope stage movements. After the completion of the tracing, each file is analyzed to determine the number and length of each axon collateral. The blind code was then broken and groups were averaged for comparison. A trace of a slide-mounted micron scale was used to convert from pixel number to microns.
Preparation of Protein Extracts.
Protein extracts from pedal ganglia were prepared as described previously (40). Briefly, ganglia were homogenized in a buffer containing 50 mM Tris⋅HCl (pH 7.8), 10 mM KCl, 0.2 mM EDTA, 0.5 mM DTT, and the protease inhibitors phenylmethylsulfonyl fluoride (1 mM), aprotinin (1 μg/ml), leupeptin (10 μg/ml), and benzamidine (5 mM). Glycerol (final concentration, 10%) was then added, followed by the addition of KCl (0.4 M final concentration). The mixture was rotated gently for 30 min and centrifuged at 14,000 × g for 30 min. The supernatant solutions were removed, dialyzed for 5 hr against ice-cold BC50 (10 mM Tris, pH 7.9/50 mM KCl/0.5 mM DTT/0.2 mM EDTA/5% glycerol) and used for electrophoretic mobility-shift assays (EMSAs) and Western blots.
EMSAs.
EMSAs were carried out by using a radioactively end-labeled, double-stranded oligonucleotide containing the CRE sequence (5′-GGCCTCCTTGGCTGACGTCAGAGAGAGAGTTCTGCA-3′ and its complementary sequence) from the rat somatostatin gene as described previously (40). The specific binding of this CRE sequence-containing probe to Aplysia CREB-like proteins has been characterized previously (19, 40, 41). The probe (0.2 ng) was incubated with equal amounts of protein extracts, as determined by a MicroBCA assay (Pierce), from control and experimental samples for 20 min at room temperature in a buffer containing 10 mM Tris, pH 7.9/5 mM MgCl2/0.5 mM EDTA/50 mM KCl/5% sucrose/5% glycerol/0.5 mM DTT/0.3 μg sonicated salmon sperm DNA/0.3 μg poly(dI-dC)(dI-dC). The oligonucleotides bound to the proteins were separated from the free oligonucleotides in a low-ionic-strength 6% polyacrylamide gel followed by autoradiography. For competition EMSAs, the cold competitors were added to the reaction mixture before the addition of protein extracts. A second CRE sequence-containing oligonucleotide from the c-fos promoter (5′-GAGTTCCGCCCAGTGACGTAGGAAGTCCATCCTGCA-3′) and an oligonucleotide containing a mutated CRE sequence but the same flanking sequences as the probe (5′-GGCCTCCTTGGCCTTAAGTGGAGAGAGAGTTCTGCA-3′) were used in these experiments. For supershift analysis, protein extracts were preincubated for 15 min with 0.5 μg of either ApCREB1 or ApCREB2 antibodies before the addition of the probe. Binding was allowed to continue for an additional 30 min in the absence of any thiol reagent. Quantification of bands for EMSAs was carried out by using a Bio-Rad model GS-670 imaging densitometer. The integrated optical densities, minus the background optical density of the film, for control and experimental samples were compared for analysis. EMSAs were repeated in at least four independent experiments.
Western Blots.
Western blotting was performed by using the same protein extracts used for the EMSAs. Equal amounts of protein extracts were resolved on an 8.5% SDS-polyacrylamide gel and transferred to an Immobilon-P membrane. A similarly prepared and loaded gel was stained with Coomassie brilliant blue to confirm equality of loading. The membrane was blocked in BSA followed by incubation with primary antibodies to either ApCREB1 or ApCREB2 (generously provided by Eric Kandel, Columbia University, Riverdale, NY) at a final concentration of 0.5 μg/ml for 3 hr. Immunoreactivity was detected by using an alkaline phosphatase-conjugated secondary antibody followed by chemiluminescence (GIBCO) as described by the vendor. Quantification of immunoreactive bands was carried out as described for EMSAs. Western blots were repeated in at least four independent experiments.
Statistical Analysis.
A Student’s t test for paired samples was used to detect differences between control and experimental groups. Raw optical densities (for EMSAs and Western blots), collateral counts, or untransformed pixel numbers (for collateral lengths) were used for this analysis. The differences were considered significant when P < 0.05. Data are presented as the mean ± SEM.
RESULTS
Axonal Injury of Tail Motor Neurons.
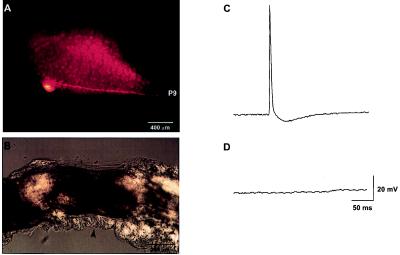
Tail motor neurons were identified visually under a microscope based on the position of their soma as described previously (42). Fig. 1A shows an example of a tail motor neuron injected with rhodamine-dextran. The pedal ganglion is visible in this photograph because of the weak autofluorescence of the cells. The axon of the injected tail motor neuron can be seen projecting down the p9 nerve. Crushing the p9 nerve severs the axons without transecting the nerve (Fig. 1B). Fig. 1C shows that stimulation of the p9 nerve using an extracellular suction electrode produced antidromic spikes, which can be recorded in the tail motor neurons. The crush blocks the generation of antidromic spikes in response to stimulation of the distal portion of p9 (Fig. 1D).
Figure 1.
(A) A representative photomicrograph indicating the location of a tail motor neuron in the pedal ganglion. The motor neuron was injected with rhodamine-dextran, and the photograph was taken under UV epifluorescent illumination. The pedal ganglion can be seen in this photograph because of the weak autofluorescence of the neurons. As reported previously by Walters et al. (42), this tail motor neuron can be seen sending its axon thorough the p9 nerve. (B) Light microscope picture of the p9 nerve showing the crush site (arrowhead). (C) Before nerve crush, antidromic spikes can be recorded in the tail motor neurons by stimulating p9 with an extracellular electrode. (D) The crush prevents antidromic spike generation when the distal end of the nerve is stimulated.
Axonal Injury Increases the Binding to a CRE Sequence-Containing Oligonucleotide.
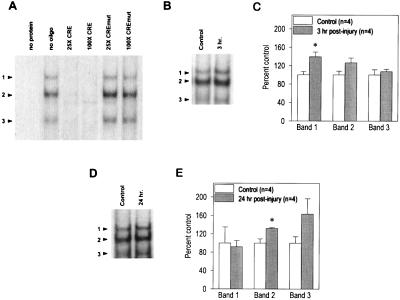
To examine whether axonal injury alters the binding of specific proteins to a CRE sequence-containing oligonucleotide, EMSAs were performed. It was difficult to obtain enough nuclear proteins from individual tail motor neurons; therefore, protein extracts from the entire pedal ganglion were utilized. Protein extracts (prepared immediately after dissection) from symmetrically located pedal ganglia were used as controls. As reported previously, Fig. 2A shows that in the presence of an Aplysia pedal ganglia extract three specifically retarded bands (arrowheads) are detected (19, 40, 41). The specific binding of the three bands was assessed by performing competition experiments. Fig. 2A shows that oligonucleotides containing the CRE sequence from the c-fos promoter (×25 and ×100 molar excess) effectively competed for binding. This oligonucleotide contains different flanking sequences than the CRE sequence-containing oligonucleotide used as the probe, indicating that the binding is a result of the presence of the CRE sequence. Consistent with this, when an oligonucleotide with a mutated CRE sequence (×25 and ×100) but identical flanking sequences to the probe was a competitor, no change in binding was observed. Fig. 2B is a picture of a representative EMSA performed by using protein extracts from control and 3-hr postinjury ganglia. The summary figure (Fig. 2C) shows the optical densities of the three bands from control and 3-hr experimental samples performed in four independent experiments. A significant increase in the optical density of only retarded band 1 was detected compared with controls (control = 100 ± 7.15% vs. 3-hr injured = 139.5 ± 10.3%, P < 0.05). By 24-hr postinjury, only the optical density of retarded band 2 was significantly elevated (Fig. 2 D and E) (control = 100 ± 9.1% vs. 24-hr injured = 131.4 ± 2.1%, P < 0.05, n = 4).
Figure 2.
Injury increases the levels of CRE-binding proteins. (A) As reported previously, pedal ganglia extracts give rise to three specifically retarded bands (arrowheads) when examined by using EMSAs (40). The specificity of the CRE sequence-containing probe for CRE-binding proteins was determined by competition EMSAs. The inclusion of a different, unlabeled CRE sequence-containing oligonucleotide effectively abolished the binding to the CRE probe at both ×25 and ×100 molar excess. In contrast, an oligonucleotide containing a mutated CRE sequence (CREmut) but identical flanking sequences to the probe did not compete for binding. (B) Picture of a representative EMSA obtained using protein extracts from control and 3-hr postinjury pedal ganglia samples. As described in Experimental Procedures, samples prepared from the uninjured symmetrical pedal ganglia were used as controls for each injured sample. (C) Summary figure showing that at 3 hr postinjury, the optical density of retarded band 1 is increased significantly as compared with the controls. The data are presented as the mean ± SEM. ∗, P < 0.05. (D) Picture of a representative EMSA obtained by using protein extracts from control and 24-hr postinjury pedal ganglia samples. (E) Summary figure indicating that at 24 hr postinjury, the optical density for the retarded band 2 is increased significantly as a result of injury. The data are presented as the mean ± SEM. ∗, P < 0.05.
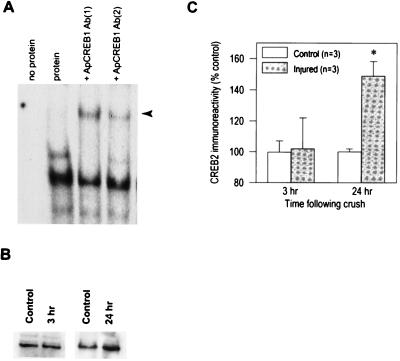
We next examined the levels of two Aplysia CRE sequence-binding proteins, ApCREB1 (an activator) and ApCREB2 (a repressor) by using Western blots and EMSAs. We were unable to detect any immunoreactivity utilizing three different antibodies to ApCREB1 possibly because of the low abundance of this protein. To establish its presence in the retarded bands we observed, supershift analysis were performed. Fig. 3A shows that when the protein extracts were preincubated with ApCREB1 antibodies, a new retarded band (arrowhead) is detected accompanied by a concurrent reduction in the optical density of retarded band 1. Similar results were obtained for each of the ApCREB1 antibodies used. When BSA was substituted for the pedal extract, no bands were detected (data not shown). Western blot analysis of ApCREB2 indicated a significant increase in ApCREB2 immunoreactivity as a result of axonal injury. Fig. 3 B and C shows that the level of ApCREB2 is enhanced at 24 hr (control = 100.0 ± 2.0% vs. 24-hr injured = 149.0 ± 9.3%, P < 0.05) but not 3 hr (control = 99.7 ± 7.3% vs. 3-hr injured = 102.0 ± 20.6) postinjury. Unfortunately, this ApCREB2 antibody failed to cause any supershift when used in EMSAs. However, it is possible that the increase we detect in retarded band 2 at 24 hr may be because of the increasing levels of ApCREB2 as a result of axonal injury.
Figure 3.
(A) Picture of a representative EMSA showing that ApCREB1 is a component of retarded band 1. When preincubated with an antibody that specifically recognizes ApCREB1, pedal extracts give rise to a new supershifted band (arrowhead) concurrent with a reduction in the intensity of retarded band 1. Similar results were obtained with each of the two different ApCREB1 antibodies utilized. (B) Picture of representative Western blots using an antibody that recognizes ApCREB2. An increase in ApCREB2 immunoreactivity can be seen in the 24-hr but not the 3-hr postinjury samples. No specific immunoreactivity was detected when the primary antibody was eliminated from the incubation mixture. (C) Summary figure showing that axonal injury significantly increases the immunoreactivity of ApCREB2 by 24 hr postinjury. The data are presented as the mean ± SEM. ∗, P < 0.05.
CRE Sequence-Containing Plasmids Can Act as TFDs.
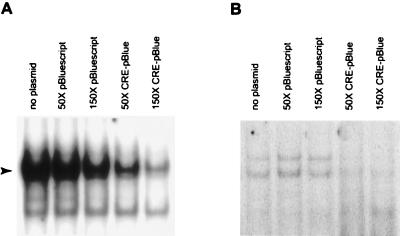
To examine the role of CRE-mediated gene expression in morphological changes associated with axonal regeneration, CRE sequence-containing plasmids were injected as TFDs to sequester CRE-binding proteins. Plasmids were used, instead of double-stranded oligonucleotides, because of their relatively longer half-lives inside the cell. The oligonucleotide containing the CRE sequence that was used as the probe for EMSA studies was subcloned into a Bluescript plasmid as described in Experimental Procedures. Before performing TFD experiments, we examined whether the CRE sequence containing plasmids could compete for binding to the CRE-binding proteins using competition EMSAs. Fig. 4 shows that plasmids lacking the CRE sequence did not change the intensity of the retarded bands by using either HeLa (Fig. 4A) or Aplysia pedal ganglia (Fig. 4B) extracts at ×50 molar excess (equivalent to 0.5 μg of plasmid DNA). At ×150 molar excess, a modest nonspecific decrease in binding was observed most likely because of the vast excess of added plasmid DNA. In contrast, CRE sequence-containing plasmids competed effectively for binding at both ×50 and ×150 molar excess.
Figure 4.
CRE sequence-containing plasmids can compete for binding of proteins. Pictures of representative EMSAs using either HeLa (A) or Aplysia pedal ganglia (B) extracts are shown. The arrowhead indicates the migration of the specifically retarded band in A. Control plasmids lacking the CRE sequence (pBluescript) do not specifically compete for binding of proteins to the radioactively labeled CRE probe. In contrast, CRE sequence-containing plasmids (CRE-pBlue) effectively compete for binding at both ×50 and ×150 molar excess.
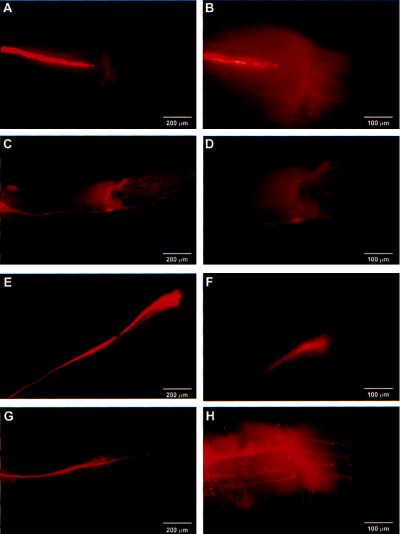
Fig. 5 shows representative morphologies of regenerating axons 1 week after the crush. Fig. 5 A and B illustrates the morphology of an axon of a tail motor neuron injected with vehicle. A few short collaterals can be seen emerging from the tip of the regenerating axon. The regenerating axon has not yet crossed the crush site, consistent with our previous studies in intact animals that showed no recovery of the tail-elicited tail–siphon withdrawal reflex at this time point (10). Fig. 5 C and D shows that injection of plasmids lacking the CRE inserts did not dramatically affect this morphology. Moreover, when a plasmid containing a TRE sequence was microinjected into the regenerating motor neuron, no obvious changes in morphology were observed as compared with controls (Fig. 5 E and F). This TRE sequence, when used as a probe in EMSA assays, specifically cross-reacts with proteins present in Aplysia CNS extracts (data not shown). In contrast to the control plasmids, microinjection of CRE sequence-containing plasmids caused numerous axon collateral formations (Fig. 5G). Fig. 5H shows a high-magnification photomicrograph of the axon shown in Fig. 5G. Similar morphological changes were detected when either 100 or 235 μg/ml concentrations of CRE sequence-containing plasmid DNA were injected.
Figure 5.
Microinjection of CRE sequence-containing plasmids as transcription factor decoys alter the morphology of regenerating motor neuron axons. The morphology of the axons was examined 1 week postinjury. Axons were visualized by injecting neurons with 2% rhodamine-dextran and photographed under UV epifluorescent illumination. (A and B) Representative pictures showing the morphology of the axon of a motor neuron injected with vehicle at two different magnifications. (C and D) Representative pictures showing the morphology of the axon of a motor neuron injected with the control plasmid lacking the CRE sequence at two different magnifications. (E and F) Representative pictures showing the morphology of the axon of a motor neuron injected with a TRE sequence-containing plasmid at two different magnifications. (G and H) Representative pictures showing the morphology of the axon of a motor neuron injected with the CRE sequence-containing plasmids at two different magnifications. Numerous collaterals can be seen emerging from the axon.
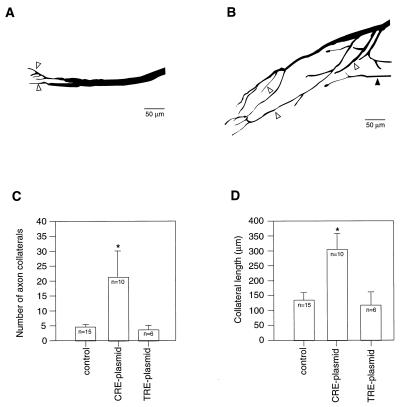
Sequestration of CRE Sequence-Binding Proteins Increases Both Axon Collateral Number and Length.
Two morphological features (number of collaterals and their lengths) were quantified by using high-resolution microscopy and neurolucida. These morphological features were examined 1 week after the crush. Representative neurolucida traces of regenerating axons from both control and CRE sequence-containing, plasmid-injected motor neurons are shown in Fig. 6A and B. Because no differences in axonal morphology between control plasmid- or saline-injected neurons were observed, these control groups were combined for further analysis. The summary of the morphological analysis is shown in Fig. 6 C and D. CRE sequence-containing plasmid injections resulted in a significant increase in the number of collaterals [control (n = 15) = 4.5 ± 0.8 vs. CRE (n = 10) = 21.2 ± 8.7, P < 0.05]. In contrast, when plasmids containing the TRE sequence were injected, no change in collateral number was detected [control = 4.5 ± 0.8 vs. TRE (n = 6) = 3.57 ± 1.06, P = 0.301]. Moreover, the average length of the collaterals also was significantly increased in the CRE sequence-containing, plasmid-injected neurons [control (n = 15) = 136.5 ± 26.5 μm vs. CRE (n = 10) = 307.1 ± 54.1 μm, P < 0.05]. The TRE plasmid-injected neurons showed no change in collateral length as compared with controls [control = 136.5 ± 26.5 μm vs. TRE (n = 6) = 123.0 ± 42.8, P = 0.646].
Figure 6.
Sequestration of CRE-binding proteins causes increased number and length of axon collaterals in regenerating motor neurons. Neurolucida traces of regenerating axons from control (A) and CRE sequence-containing, plasmid-injected (B) neurons. Collaterals can be seen emerging from the regenerating axon (e.g., open arrowhead). In the CRE sequence-containing, plasmid-injected cells, collaterals emerged not only from the tip, but also from the body of the main axon. Some collaterals (solid arrowhead) emerge from the main axon outside the shown field of view. (C) Summary figure showing the average number of collaterals in control and TRE sequence- and CRE sequence-containing, plasmid-injected neurons. The data are presented as the mean ± SEM. ∗, P < 0.05. (D) Summary figure showing the average length of axon collaterals in the control and TRE sequence- and CRE sequence-containing, plasmid-injected neurons. The data are presented as the mean ± SEM. ∗, P < 0.05.
DISCUSSION
Intrinsic genetic programs, possibly influenced by environmental factors, have been hypothesized to control regeneration in the mature CNS (2). However, the identities of the genomic programs that contribute to axonal regeneration remain to be determined. In this study, we demonstrate that CRE-binding proteins (both activators and repressors) contribute to axon collateral elaboration in regenerating Aplysia tail motor neurons. Microinjection of CRE sequence-containing plasmids to sequester CRE-binding proteins caused robust collateral sprouting of motor neuron axons. Collateral formation is thought to be an important component of axonal regeneration as well as guidance during CNS development (3, 4, 43). Trophic factors such as nerve growth factor, brain-derived neurotrophic factor, and neurotrophin-3 (NT)-3 have been shown to induce collateral formation whereas axonal guidance has been shown to be facilitated by molecules such as netrins. For example, injection of NT-3 enhances collateral sprouting in the corticospinal tract after axonal injury (44). Binding of these trophic factors to their plasma membrane receptors activates the MAPK pathway. MAPK has been shown to activate p90rsk, which, in turn, can stimulate transcription of CRE sequence-containing genes via CREB phosphorylation (17, 45). We propose that modulation of a genetic program via CREB and related proteins is of importance for regeneration in the adult CNS and possibly axonal guidance during development.
The activity of CRE-binding activators and repressors can be modulated by phosphorylation and/or enhanced expression (29, 30). To examine the expression of CRE-binding proteins after axonal injury, we performed EMSAs. We detected increased levels of CRE-binding proteins at both 3 and 24 hr postinjury (Fig. 2). At the 3-hr time point, the optical density of retarded band 1 was significantly increased whereas at the 24-hr time point, the optical density of retarded band 2 was increased significantly compared with controls (Fig. 2 C and E). Although not statistically significant, retarded band 3 also showed a modest increase as a result of axonal injury at the 24-hr time point. These results suggest that the pattern of expression of CRE-binding proteins changes over time as a result of injury. To date, only one activator (ApCREB1) and one repressor (ApCREB2) have been isolated and characterized in Aplysia CNS (21). Using antibodies against these proteins, supershift EMSA analysis was performed to identify the constituents of the retarded bands. Consistent with our previously published findings using an antibody raised against mammalian CREB (41), it was found that ApCREB1 is a component of retarded band 1. Although ApCREB1 was identified in retarded band 1, and this band was found to be increased significantly at the 3-hr time point, we were unable to determine independently whether ApCREB1 levels are indeed changing as a result of axonal injury. The composition of the other two retarded bands is unknown at present.
It has been hypothesized that the relative activities of activators to repressors determine the ensuing induction or repression of CRE sequence-containing target genes and the subsequent morphological changes (21–23, 26, 46). In Aplysia, injection of neutralizing antibodies to the repressor ApCREB2 has been reported to facilitate the growth of new synaptic connections in response to serotonin exposure (21). This finding suggests that decreased activity of CRE-binding repressors (which would result in an increased relative activity of activators to repressors) enhances morphological changes. A recent study showed that decreasing the level of CREB protein by antisense oligonucleotides blocks spine formation in hippocampal pyramidal neurons in response to estradiol exposure, further supporting the idea that altering the ratio of activators to repressors affects neuronal morphology (47). We demonstrated that injection of CRE sequence-containing plasmids caused collateral elaboration (increased number and length) in regenerating tail motor neuron axons (Figs. 5 and 6). The biochemical characterizations of ApCREB1 and ApCREB2 indicate that ApCREB1 has a higher affinity for the CRE sequence as compared with ApCREB2 (21). These binding affinities were shown to be unaffected by phosphorylation in response to transmitter application. This suggests that ApCREB1, and perhaps other related activators, is bound tightly whereas repressors are bound weakly to the promoters of CRE sequence-containing genes in the motor neurons in vivo. This may result in preferential sequestration of repressors over activators. In addition, our Western blot analysis revealed that the level of ApCREB2 is significantly enhanced as a result of axonal injury. If repressors such as ApCREB2 are being synthesized as a result of injury, they could be readily sequestered by TFDs before reaching their genomic targets. Although other interpretations are plausible, we suggest that the CRE sequence-containing TFDs might be sequestering CRE repressors more effectively than activators in these regenerating motor neurons, resulting in enhanced target gene expression and increased collateral formation.
Recent studies have shown that CRE-binding proteins (both activators and repressors) participate in long-term memory formation as well as the associated morphological changes (21, 22, 26, 47, 48). For example, in Aplysia sensory neurons, injection of cAMP or blocking the CRE-binding repressor ApCREB2 increases the number of branch points, varicosities, and synapses (21, 39). The findings presented in this report are consistent with a role for CRE-mediated gene expression in morphological changes and further suggest that the balance between CRE-binding activator and repressor functions is critical for regeneration of motor neuron axons. Specifically, our results indicate that axonal injury results in increased levels of CRE sequence-binding repressors, presumably leading to decreased expression of CRE sequence-containing genes. The reason for suppression of CRE-mediated gene expression is not clear at present. It would be interesting to examine whether the sequestration of CRE-binding proteins induces collateral elaboration in uninjured motor neurons. Unfortunately, it is difficult to assess this effect in our preparation because the removal of the ganglia transects all the nerves and most likely injures all motor neurons. Although technically demanding, a semi-intact or intact preparation could be used to test this possibly in the future.
Acknowledgments
The helpful comments of Drs. William Frost, Frank Adams, and Leonard Cleary are gratefully acknowledged. The authors also thank Rama Grenda and Marcy Wainwright for their help with the figures and neurolucida, respectively. The authors thank Dr. Eric Kandel for his generous gift of the Aplysia CREB1 and CREB2 antibodies. This work was supported by grants from the National Institutes of Health (MH49662 and NS35457).
ABBREVIATIONS
- CRE
cAMP response element
- CNS
central nervous system
- CREB
CRE-binding protein
- TFD
transcription factor decoy
- EMSA
electrophoretic mobility-shift assay
References
- 1.Aguayo A J, Rasminsky M, Bray G M, Carbonetto S, McKerracher L, Villegas-Perez M P, Vidal-Sanz M, Carter D A. Philos Trans Roy Soc London. 1991;331:337–343. doi: 10.1098/rstb.1991.0025. [DOI] [PubMed] [Google Scholar]
- 2.Chen D F, Jhaveri S, Schneider G E. Proc Natl Acad Sci USA. 1995;92:7287–7291. doi: 10.1073/pnas.92.16.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jhaveri S, Edwards M A, Schneider G E. Exp Brain Res. 1991;87:371–382. doi: 10.1007/BF00231854. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy T E, Tessier-Lavigne M. Curr Opin Neurobiol. 1995;5:83–90. doi: 10.1016/0959-4388(95)80091-3. [DOI] [PubMed] [Google Scholar]
- 5.Ambron R T, Walters E T. Mol Neurobiol. 1996;13:61–79. doi: 10.1007/BF02740752. [DOI] [PubMed] [Google Scholar]
- 6.Buriani A, Savage M J, Burmeister D W, Goldberg D J. J Neurochem. 1990;55:1817–1820. doi: 10.1111/j.1471-4159.1990.tb04975.x. [DOI] [PubMed] [Google Scholar]
- 7.Skene J H P. Annu Rev Neurosci. 1989;12:127–156. doi: 10.1146/annurev.ne.12.030189.001015. [DOI] [PubMed] [Google Scholar]
- 8.Savage M J, Buriani A, Goldberg D J. J Neurochem. 1990;54:270–276. doi: 10.1111/j.1471-4159.1990.tb04912.x. [DOI] [PubMed] [Google Scholar]
- 9.Koo E H, Hoffman P N, Price D L. Brain Res. 1988;449:361–363. doi: 10.1016/0006-8993(88)91054-2. [DOI] [PubMed] [Google Scholar]
- 10.Noel F, Frost W N, Tian L-M, Colicos M A, Dash P K. J Neurosci. 1995;15:6926–6938. doi: 10.1523/JNEUROSCI.15-10-06926.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oblinger M M, Szumlas R A, Wong J, Liuzzi F J. J Neurosci. 1989;9:2645–2653. doi: 10.1523/JNEUROSCI.09-08-02645.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziv N E, Spira M E. Eur J Neurosci. 1993;5:657–668. doi: 10.1111/j.1460-9568.1993.tb00531.x. [DOI] [PubMed] [Google Scholar]
- 13.Abrams T W, Karl K A, Kandel E R. J Neurosci. 1991;11:2655–2665. doi: 10.1523/JNEUROSCI.11-09-02655.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Byrne J H, Zwartjes R, Homayouni R, Critz S S, Eskin A. Second Messenger Phosphoprotein Res. 1993;27:47–108. [PubMed] [Google Scholar]
- 15.Hanson P I, Schulman H. Annu Rev Biochem. 1992;61:559–601. doi: 10.1146/annurev.bi.61.070192.003015. [DOI] [PubMed] [Google Scholar]
- 16.Hawkins R D, Kandel E R, Siegelbaum S A. Annu Rev Neurosci. 1993;16:625–665. doi: 10.1146/annurev.ne.16.030193.003205. [DOI] [PubMed] [Google Scholar]
- 17.Xing J, Ginty D D, Greenberg M E. Science. 1996;270:1326–1331. [Google Scholar]
- 18.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 19.Alberinni C M, Ghirardi M, Metz R, Kandel E R. Cell. 1994;75:1263–1271. doi: 10.1016/0092-8674(94)90386-7. [DOI] [PubMed] [Google Scholar]
- 20.Bailey C H, Bartsch D, Kandel E R. Proc Natl Acad Sci USA. 1996;93:13445–13452. doi: 10.1073/pnas.93.24.13445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartsch D, Ghirardi M, Skehel P A, Karl K A, Herder S P, Chen M, Bailey C H, Kandel E R. Cell. 1995;83:979–992. doi: 10.1016/0092-8674(95)90213-9. [DOI] [PubMed] [Google Scholar]
- 22.Carew T J. Neuron. 1996;16:5–8. doi: 10.1016/s0896-6273(00)80016-1. [DOI] [PubMed] [Google Scholar]
- 23.Martin K C, Kandel E R. Neuron. 1996;17:567–570. doi: 10.1016/s0896-6273(00)80188-9. [DOI] [PubMed] [Google Scholar]
- 24.O’Leary F A, Byrne J H, Cleary L J. J Neurosci. 1995;15:3519–3525. doi: 10.1523/JNEUROSCI.15-05-03519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schacher S, Kandel E R, Montarolo P. Neuron. 1993;10:1079–1088. doi: 10.1016/0896-6273(93)90056-w. [DOI] [PubMed] [Google Scholar]
- 26.Yin J C P, Del Vecchio M, Zhou H, Tully T. Drosophila Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 27.Montminy M R, Sevarino K A, Wagner J A, Mandel G, Goodman R H. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng M, Greenberg M E. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez G A, Yamamoto K A, Fisher W H, Karr D, Menzel P, Bigg W, III, Vale W W, Montminy M R. Nature (London) 1989;337:749–752. doi: 10.1038/337749a0. [DOI] [PubMed] [Google Scholar]
- 30.Lalli E, Sassone-Corsi P. J Biol Chem. 1994;269:17359–17362. [PubMed] [Google Scholar]
- 31.Dash P K, Karl K A, Colicos M A, Prywes R, Kandel E R. Proc Natl Acad Sci USA. 1991;88:5061–5063. doi: 10.1073/pnas.88.11.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ginty D D, Kornhauser J M, Thompson M A, Bading H, Mayo K E, Takahashi J S, Greenberg M E. Science. 1993;260:238–241. doi: 10.1126/science.8097062. [DOI] [PubMed] [Google Scholar]
- 33.Sheng M, Thomson M A, Greenberg M E. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- 34.Sun P, Enslen H, Myung P S, Maurer R A. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- 35.Bartsch D, Casadio A, Karl K, Serodio P, Kandel E R. Soc Neurosci Abstr. 1997;3:233. [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 37.Coggeshall R E, Yaksta B A, Swartz F J. Chromosoma. 1971;32:205–212. doi: 10.1007/BF00286009. [DOI] [PubMed] [Google Scholar]
- 38.Lasek R J, Dower W J. Science. 1971;172:278–280. doi: 10.1126/science.172.3980.278. [DOI] [PubMed] [Google Scholar]
- 39.Nazif F A, Byrne J H, Cleary L J. Brain Res. 1991;539:324–327. doi: 10.1016/0006-8993(91)91638-h. [DOI] [PubMed] [Google Scholar]
- 40.Dash P K, Hochner B, Kandel E R. Nature (London) 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 41.Dash P K, Moore A N. Mol Brain Res. 1996;39:43–51. doi: 10.1016/0169-328x(95)00350-2. [DOI] [PubMed] [Google Scholar]
- 42.Walters E T, Byrne J H, Carew T J, Kandel E R. J Neurophys. 1983;50:1522–1542. doi: 10.1152/jn.1983.50.6.1522. [DOI] [PubMed] [Google Scholar]
- 43.Henderson Z. Prog Neurobiol. 1996;48:219–254. doi: 10.1016/0301-0082(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 44.Schnell L, Schneider R, Kolbeck R, Barde Y A, Schwab M E. Nature (London) 1994;367:170–173. doi: 10.1038/367170a0. [DOI] [PubMed] [Google Scholar]
- 45.Sturgill T W, Ray L B, Erikson E, Maller J L. Nature (London) 1988;334:715–718. doi: 10.1038/334715a0. [DOI] [PubMed] [Google Scholar]
- 46.Bailey C H, Alberini C, Ghirardi M, Kandel E R. Adv Second Messenger Phosphoprotein Res. 1994;29:529–544. doi: 10.1016/s1040-7952(06)80034-0. [DOI] [PubMed] [Google Scholar]
- 47.Murphy D D, Segal M. Proc Natl Acad Sci USA. 1997;94:1482–1487. doi: 10.1073/pnas.94.4.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guzowski J F, McGaugh J L. Proc Natl Acad Sci USA. 1997;94:2693–2698. doi: 10.1073/pnas.94.6.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molina C A, Foulkes N S, Lalli E, Sassone-Corsi P. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]