Abstract
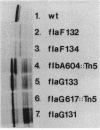
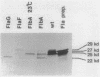
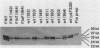
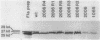
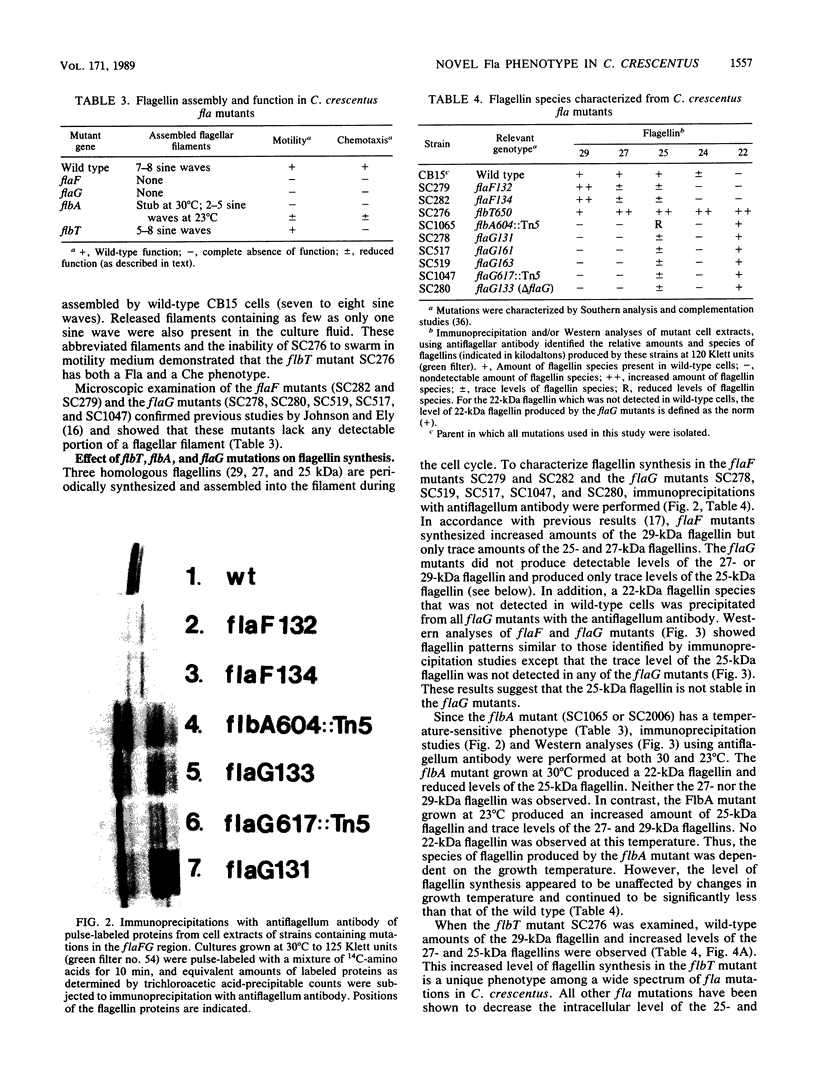
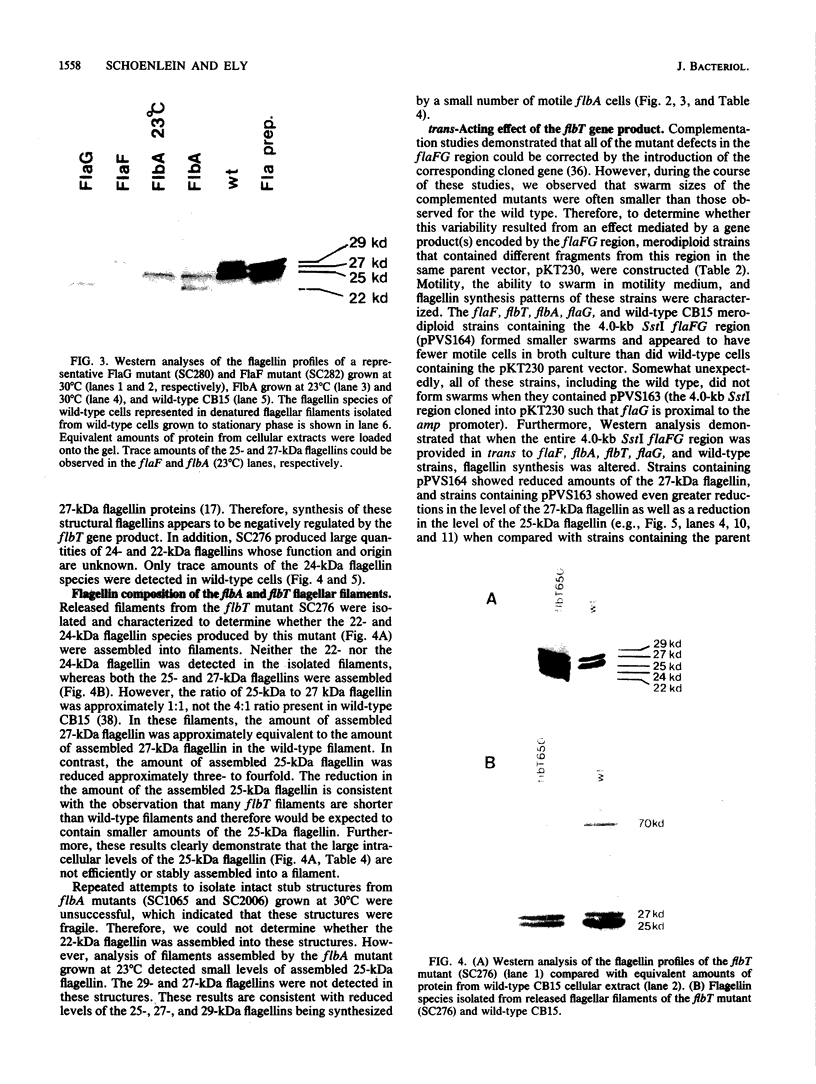
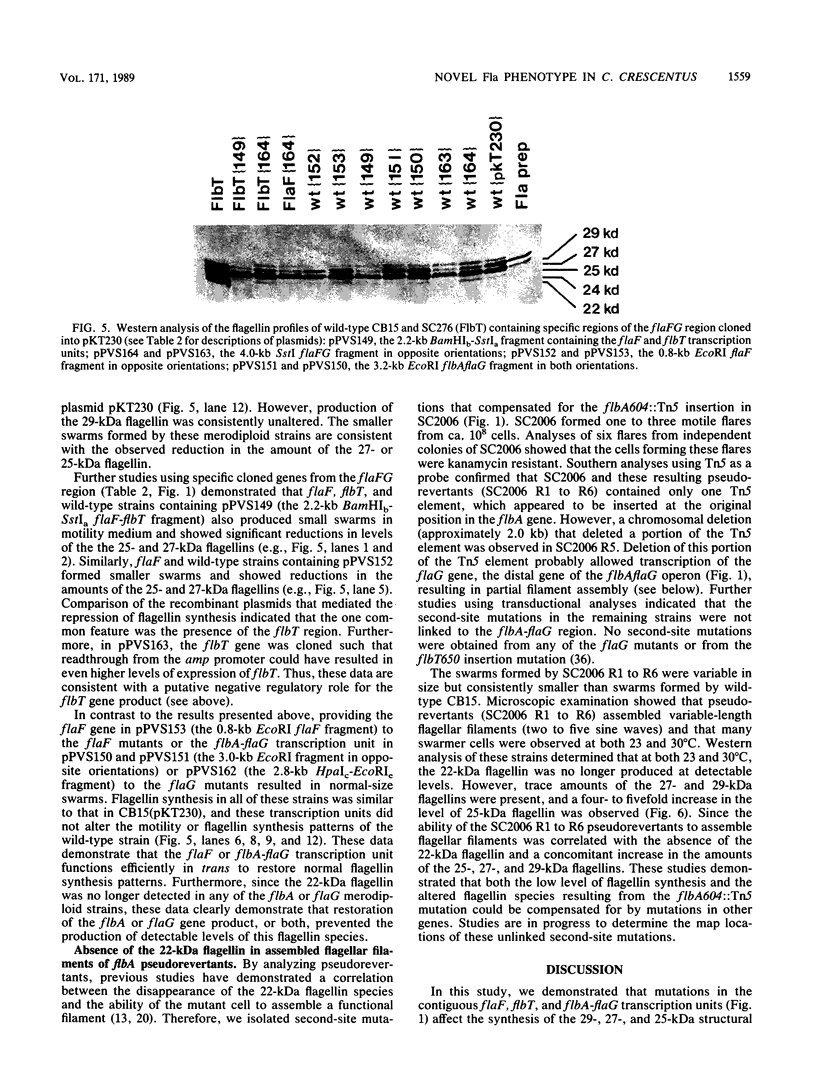
During the Caulobacter crescentus cell cycle, flagellin synthesis and filament assembly are temporally controlled events which require the products encoded by the contiguous flaF, flbT, and flbA-flaG transcription units (P.V. Schoenlein, L.S. Gallman, and B. Ely, J. Bacteriol. 171:000-000, 1989). To better define the functions of these genes, immunoprecipitation studies, Western blot (immunoblot) analyses, and electron microscopic analyses characterized flagellin synthesis and assembly in mutant and merodiploid strains. Mutations in the flaF or flbA-flaG transcription unit resulted in reduced synthesis of the 25- and 27-kilodalton (kDa) flagellins. In contrast, mutations in flbT resulted in overproduction of these flagellins. The FlbT phenotype is unique, since all other identified C. crescentus fla mutations cause a reduction in the levels of the 25- and 27-kDa flagellins. Furthermore, the flbT mutant showed a chemotaxis deficiency even though it was motile. Thus, the flbT gene product appears to be involved in the regulation of both flagellin synthesis and chemotactic function. Mutations in the flbT and flbA-flaG transcription units also resulted in the production of a 22-kDa flagellin species that is not normally detected in wild-type cells. This flagellin species was not detected in the flbT filaments. Furthermore, the 22-kDa flagellin was no longer detected in flbA pseudorevertants that assembled functional filaments. Thus, the 22-kDa flagellin does not appear to be assembled into filaments. Since many of the flbT filaments are shorter than wild-type filaments, we discuss the possibility that the 22-kDa flagellin species may adversely affect flagellin assembly in this mutant.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bryan R., Champer R., Gomes S., Ely B., Shapiro L. Separation of temporal control and trans-acting modulation of flagellin and chemotaxis genes in Caulobacter. Mol Gen Genet. 1987 Feb;206(2):300–306. doi: 10.1007/BF00333588. [DOI] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- Chen L. S., Mullin D., Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci U S A. 1986 May;83(9):2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H., Gerardot C. J. Genetic mapping of genes required for motility in Caulobacter crescentus. Genetics. 1984 Nov;108(3):523–532. doi: 10.1093/genetics/108.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J., Fleming D. L., Gomes S. L., Frederikse P., Shapiro L. General nonchemotactic mutants of Caulobacter crescentus. Genetics. 1986 Nov;114(3):717–730. doi: 10.1093/genetics/114.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Johnson R. C. Generalized Transduction in CAULOBACTER CRESCENTUS. Genetics. 1977 Nov;87(3):391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Vectors for transposon mutagenesis of non-enteric bacteria. Mol Gen Genet. 1985;200(2):302–304. doi: 10.1007/BF00425440. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda A., Asada M., Koyasu S., Yoshida H., Yaginuma K., Okada Y. Regulation of polar morphogenesis in Caulobacter crescentus. J Bacteriol. 1981 Jan;145(1):559–572. doi: 10.1128/jb.145.1.559-572.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P. R., Agabian N. The nucleotide sequence of the Mr = 28,500 flagellin gene of Caulobacter crescentus. J Biol Chem. 1983 Jun 25;258(12):7395–7401. [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Isolation of spontaneously derived mutants of Caulobacter crescentus. Genetics. 1977 May;86(1):25–32. doi: 10.1093/genetics/86.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ferber D. M., Ely B. Synthesis and assembly of flagellar components by Caulobacter crescentus motility mutants. J Bacteriol. 1983 Jun;154(3):1137–1144. doi: 10.1128/jb.154.3.1137-1144.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Walsh M. P., Ely B., Shapiro L. Flagellar hook and basal complex of Caulobacter crescentus. J Bacteriol. 1979 Jun;138(3):984–989. doi: 10.1128/jb.138.3.984-989.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S., Asada M., Fukuda A., Okada Y. Sequential polymerization of flagellin A and flagellin B into Caulobacter flagella. J Mol Biol. 1981 Dec 5;153(2):471–475. doi: 10.1016/0022-2836(81)90292-8. [DOI] [PubMed] [Google Scholar]
- Koyasu S. On flagellar formation in Caulobacter crescentus: novel flagellin synthesis in stub-forming non-motile mutants of C. crescentus. J Biochem. 1984 Nov;96(5):1351–1364. doi: 10.1093/oxfordjournals.jbchem.a134963. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lagenaur C., Agabian N. Physical characterization of Caulobacter crescentus flagella. J Bacteriol. 1976 Oct;128(1):435–444. doi: 10.1128/jb.128.1.435-444.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy Z. G., Bryan R. A., Reuter S. H., Shapiro L. Control of synthesis and positioning of a Caulobacter crescentus flagellar protein. Genes Dev. 1987 Aug;1(6):626–635. doi: 10.1101/gad.1.6.626. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983 Apr 14;302(5909):630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- Minnich S. A., Newton A. Promoter mapping and cell cycle regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Swanson E., Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985 Nov 5;186(1):107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- Ohta N., Swanson E., Ely B., Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984 Jun;158(3):897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Newton A. Temporal control of the cell cycle in Caulobacter crescentus: roles of DNA chain elongation and completion. J Mol Biol. 1980 Mar 25;138(1):109–128. doi: 10.1016/s0022-2836(80)80007-6. [DOI] [PubMed] [Google Scholar]
- Osley M. A., Sheffery M., Newton A. Regulation of flagellin synthesis in the cell cycle of caulobacter: dependence on DNA replication. Cell. 1977 Oct;12(2):393–400. doi: 10.1016/0092-8674(77)90115-5. [DOI] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purucker M., Bryan R., Amemiya K., Ely B., Shapiro L. Isolation of a Caulobacter gene cluster specifying flagellum production by using nonmotile Tn5 insertion mutants. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6797–6801. doi: 10.1073/pnas.79.22.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., Henderson N. The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene. 1988 Oct 30;70(2):331–341. doi: 10.1016/0378-1119(88)90205-3. [DOI] [PubMed] [Google Scholar]
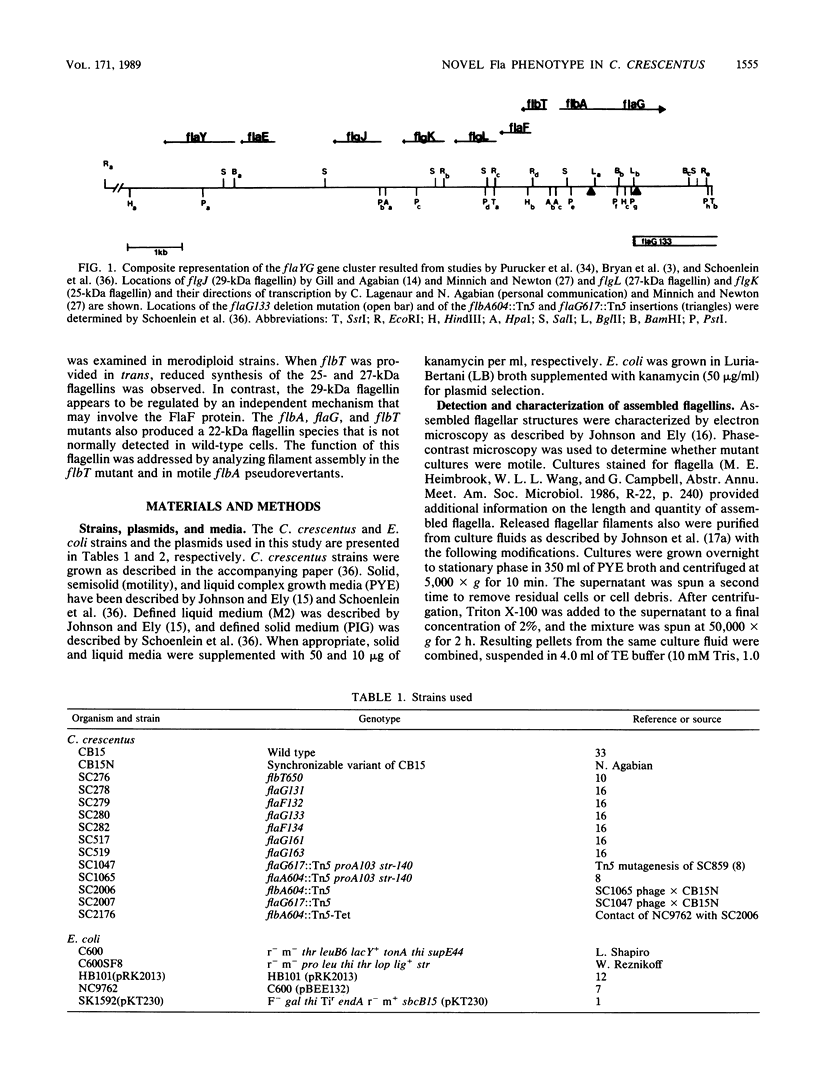
- Schoenlein P. V., Gallman L. S., Ely B. Organization of the flaFG gene cluster and identification of two additional genes involved in flagellum biogenesis in Caulobacter crescentus. J Bacteriol. 1989 Mar;171(3):1544–1553. doi: 10.1128/jb.171.3.1544-1553.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Regulation of periodic protein synthesis in the cell cycle: control of initiation and termination of flagellar gene expression. Cell. 1981 Apr;24(1):49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- Weissborn A., Steinmann H. M., Shapiro L. Characterization of the proteins of the Caulobacter crescentus flagellar filament. Peptide analysis and filament organization. J Biol Chem. 1982 Feb 25;257(4):2066–2074. [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Ishihara A., Aizawa S., Macnab R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986 Apr;166(1):187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]