Abstract
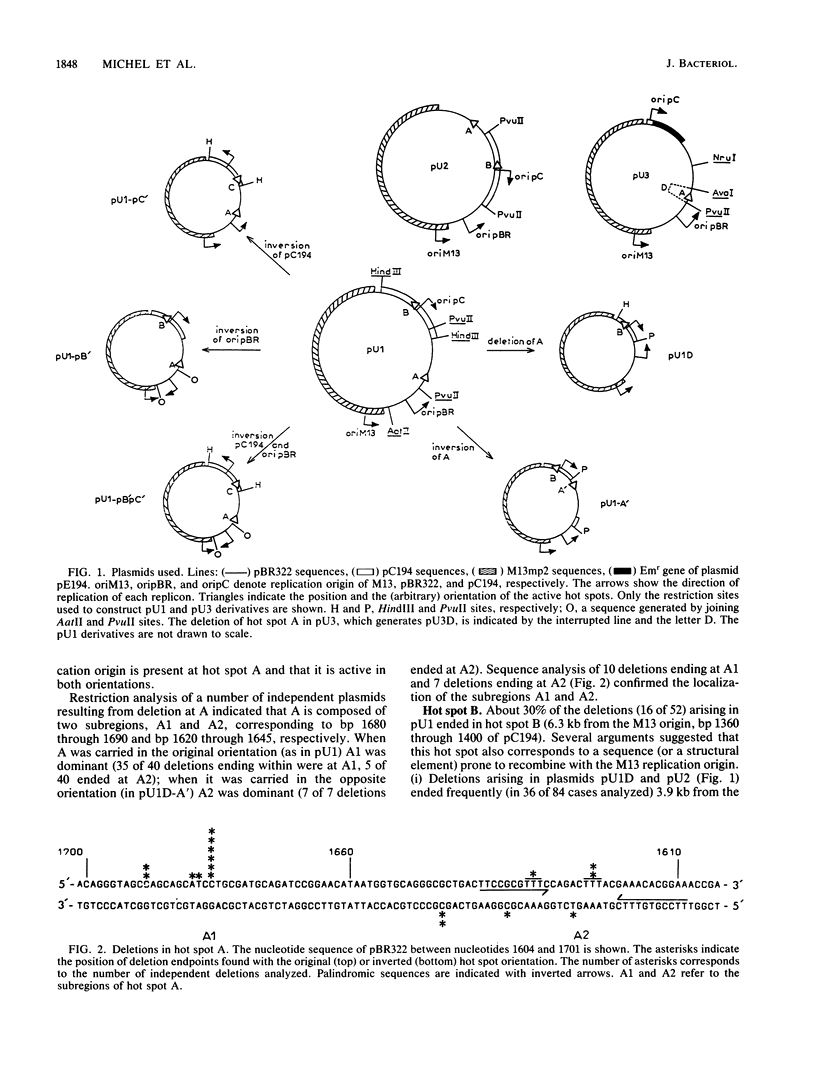
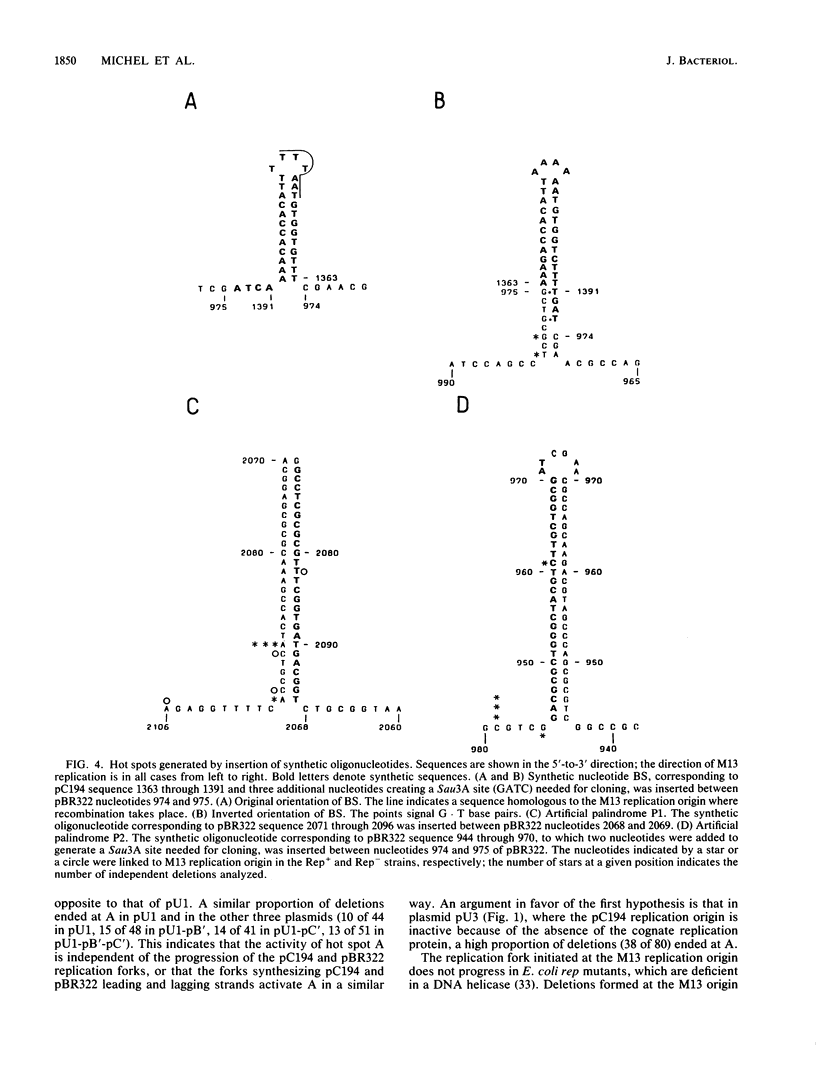
Deletions form frequently in chimeric plasmids composed of M13mp2, pBR322, and pC194 (B. Michel and S. D. Ehrlich, Proc. Natl. Acad. Sci. USA 83:3386-3390, 1986). They are generated by joining of the nucleotide neighboring the nick site in the M13 replication origin to a nonadjacent nucleotide. This nucleotide is most often located within particular short plasmid regions, named deletion hot spots. Three natural hot spots were present in the chimeric plasmids. Two were active only when the DNA replication initiated at the M13 origin was allowed to progress; the third was active only in the presence of wild-type amounts of DNA ligase. Three artificial hot spots were generated by creating palindromic sequences in the plasmids.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Anderson P. Anecdotal, historical and critical commentaries on genetics twenty years of illegitimate recombination. Genetics. 1987 Apr;115(4):581–583. doi: 10.1093/genetics/115.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backman K., Boyer H. W. Tetracycline resistance determined by pBR322 is mediated by one polypeptide. Gene. 1983 Dec;26(2-3):197–203. doi: 10.1016/0378-1119(83)90190-7. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brunier D., Michel B., Ehrlich S. D. Copy choice illegitimate DNA recombination. Cell. 1988 Mar 25;52(6):883–892. doi: 10.1016/0092-8674(88)90430-8. [DOI] [PubMed] [Google Scholar]
- Bullock P., Champoux J. J., Botchan M. Association of crossover points with topoisomerase I cleavage sites: a model for nonhomologous recombination. Science. 1985 Nov 22;230(4728):954–958. doi: 10.1126/science.2997924. [DOI] [PubMed] [Google Scholar]
- Colman A., Byers M. J., Primrose S. B., Lyons A. Rapid purification of plasmid DNAs by hydroxyapatite chromatography. Eur J Biochem. 1978 Nov 2;91(1):303–310. doi: 10.1111/j.1432-1033.1978.tb20966.x. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Ehrlich S. D. DNA cloning in Bacillus subtilis. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1433–1436. doi: 10.1073/pnas.75.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Gottesman M. M., Hicks M. L., Gellert M. Genetics and function of DNA ligase in Escherichia coli. J Mol Biol. 1973 Jul 15;77(4):531–547. doi: 10.1016/0022-2836(73)90221-0. [DOI] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi K., Ravetch J. V., Zinder N. D. DNA replication of bacteriophage f1 in vivo. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):389–399. doi: 10.1101/sqb.1979.043.01.045. [DOI] [PubMed] [Google Scholar]
- Horowitz B., Deonier R. C. Formation of delta tra F' plasmids: specific recombination at oriT. J Mol Biol. 1985 Nov 20;186(2):267–274. doi: 10.1016/0022-2836(85)90103-2. [DOI] [PubMed] [Google Scholar]
- Huang C. C., Hearst J. E., Alberts B. M. Two types of replication proteins increase the rate at which T4 DNA polymerase traverses the helical regions in a single-stranded DNA template. J Biol Chem. 1981 Apr 25;256(8):4087–4094. [PubMed] [Google Scholar]
- Huang C. C., Hearst J. E. Pauses at positions of secondary structure during in vitro replication of single-stranded fd bacteriophage DNA by T4 DNA polymerase. Anal Biochem. 1980 Mar 15;103(1):127–139. doi: 10.1016/0003-2697(80)90246-8. [DOI] [PubMed] [Google Scholar]
- Ikeda H. Bacteriophage T4 DNA topoisomerase mediates illegitimate recombination in vitro. Proc Natl Acad Sci U S A. 1986 Feb;83(4):922–926. doi: 10.1073/pnas.83.4.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Moriya K., Matsumoto T. In vitro study of illegitimate recombination: involvement of DNA gyrase. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):399–408. doi: 10.1101/sqb.1981.045.01.054. [DOI] [PubMed] [Google Scholar]
- Iordănescu S. Recombinant plasmid obtained from two different, compatible staphylococcal plasmids. J Bacteriol. 1975 Nov;124(2):597–601. doi: 10.1128/jb.124.2.597-601.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jannière L., Ehrlich S. D. Recombination between short repeated sequences is more frequent in plasmids than in the chromosome of Bacillus subtilis. Mol Gen Genet. 1987 Nov;210(1):116–121. doi: 10.1007/BF00337766. [DOI] [PubMed] [Google Scholar]
- Kaguni L. S., Clayton D. A. Template-directed pausing in in vitro DNA synthesis by DNA polymerase a from Drosophila melanogaster embryos. Proc Natl Acad Sci U S A. 1982 Feb;79(4):983–987. doi: 10.1073/pnas.79.4.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Jensch F., von Depka-Prondzynski M., Fritz H. J., Borgmeyer U., Mizuuchi K. Resolution of Holliday structures by endonuclease VII as observed in interactions with cruciform DNA. Cold Spring Harb Symp Quant Biol. 1984;49:815–825. doi: 10.1101/sqb.1984.049.01.092. [DOI] [PubMed] [Google Scholar]
- König H., Riedel H. D., Knippers R. Reactions in vitro of the DNA polymerase-primase from Xenopus laevis eggs. A role for ATP in chain elongation. Eur J Biochem. 1983 Oct 3;135(3):435–442. doi: 10.1111/j.1432-1033.1983.tb07670.x. [DOI] [PubMed] [Google Scholar]
- LaDuca R. J., Fay P. J., Chuang C., McHenry C. S., Bambara R. A. Site-specific pausing of deoxyribonucleic acid synthesis catalyzed by four forms of Escherichia coli DNA polymerase III. Biochemistry. 1983 Oct 25;22(22):5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- Lane H. E., Denhardt D. T. The rep mutation. III. Altered structure of the replicating Escherichia coli chromosome. J Bacteriol. 1974 Nov;120(2):805–814. doi: 10.1128/jb.120.2.805-814.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D. R., Stahl F. W. Viability of lambda phages carrying a perfect palindrome in the absence of recombination nucleases. 1983 Sep 29-Oct 5Nature. 305(5933):448–451. doi: 10.1038/305448a0. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Ehrlich S. D. In vitro genetic labeling of Bacillus subtilis cryptic plasmid pHV400. Plasmid. 1979 Jan;2(1):48–58. doi: 10.1016/0147-619x(79)90005-2. [DOI] [PubMed] [Google Scholar]
- Primrose S. B., Ehrlich S. D. Isolation of plasmid deletion Mutants and study of their instability. Plasmid. 1981 Sep;6(2):193–201. doi: 10.1016/0147-619x(81)90066-4. [DOI] [PubMed] [Google Scholar]
- Reckmann B., Grosse F., Urbanke C., Frank R., Blöcker H., Krauss G. Analysis of secondary structures in M13mp8 (+) single-stranded DNA by the pausing of DNA polymerase alpha. Eur J Biochem. 1985 Nov 4;152(3):633–643. doi: 10.1111/j.1432-1033.1985.tb09242.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller H. The intergenic region and the origins for filamentous phage DNA replication. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):401–408. doi: 10.1101/sqb.1979.043.01.046. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver D. T., DePamphilis M. L. The role of palindromic and non-palindromic sequences in arresting DNA synthesis in vitro and in vivo. J Mol Biol. 1984 Dec 25;180(4):961–986. doi: 10.1016/0022-2836(84)90266-3. [DOI] [PubMed] [Google Scholar]
- de Massy B., Weisberg R. A., Studier F. W. Gene 3 endonuclease of bacteriophage T7 resolves conformationally branched structures in double-stranded DNA. J Mol Biol. 1987 Jan 20;193(2):359–376. doi: 10.1016/0022-2836(87)90224-5. [DOI] [PubMed] [Google Scholar]
- van Wezenbeek P. M., Hulsebos T. J., Schoenmakers J. G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980 Oct;11(1-2):129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]


