Abstract
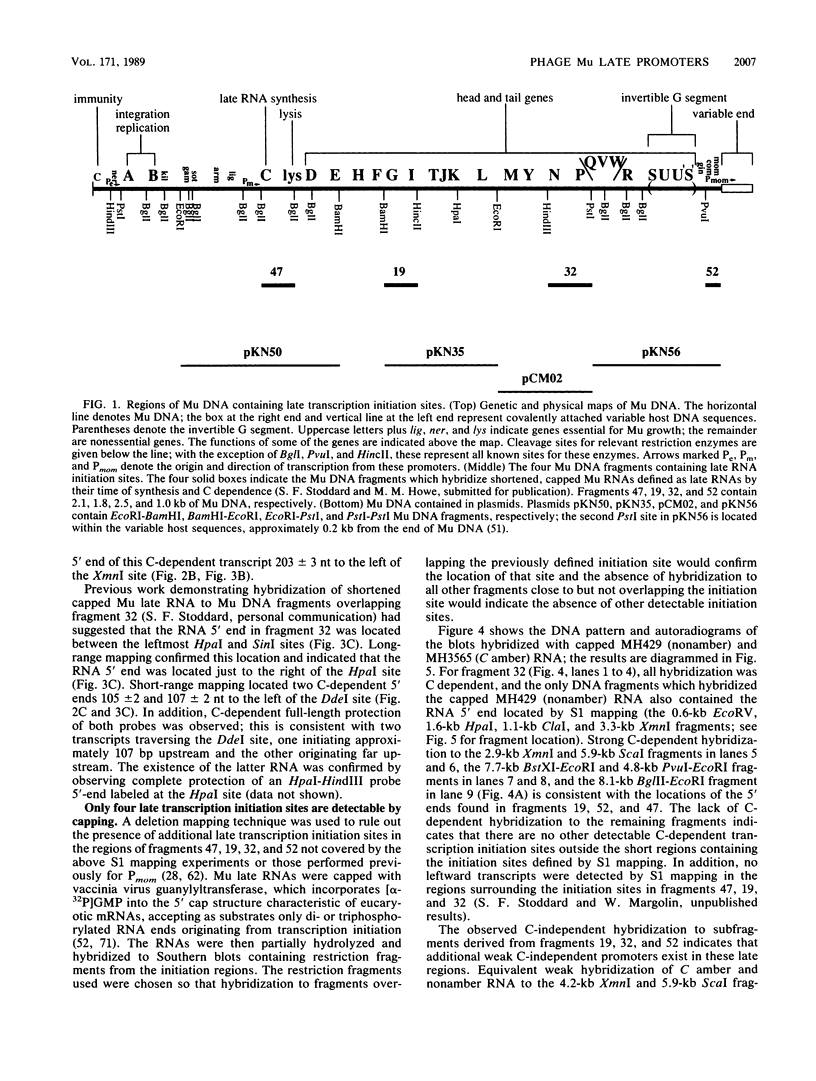
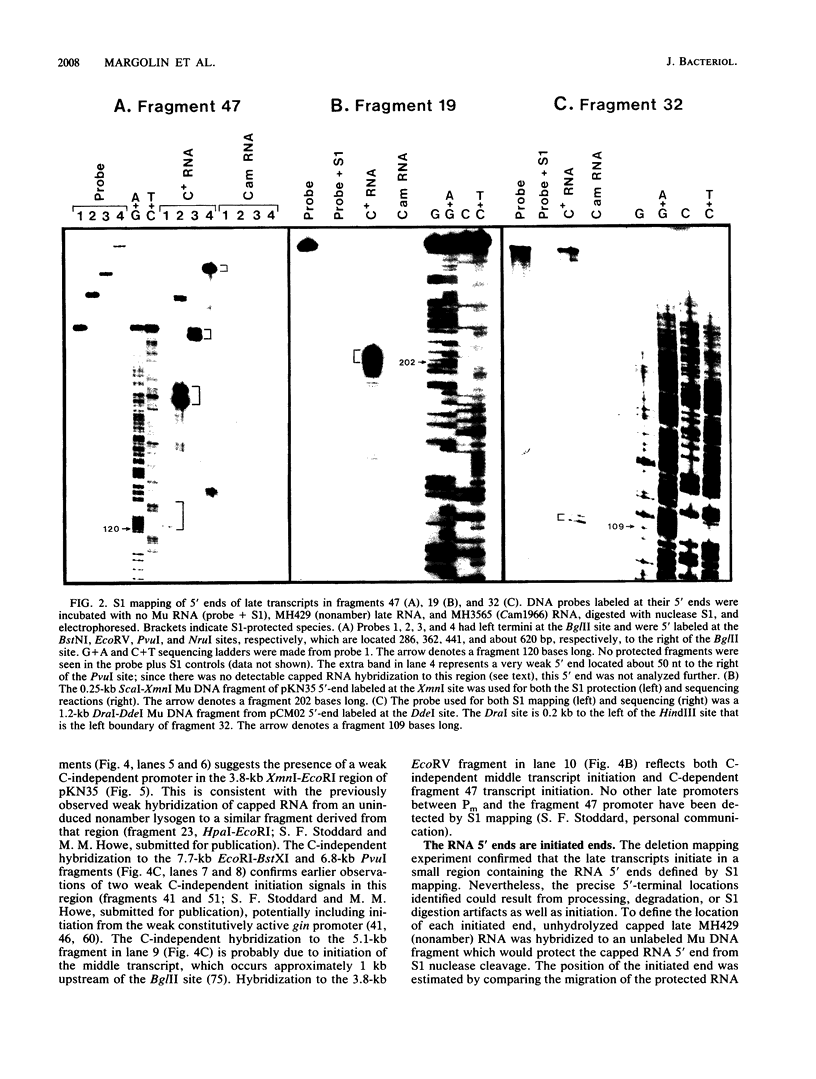
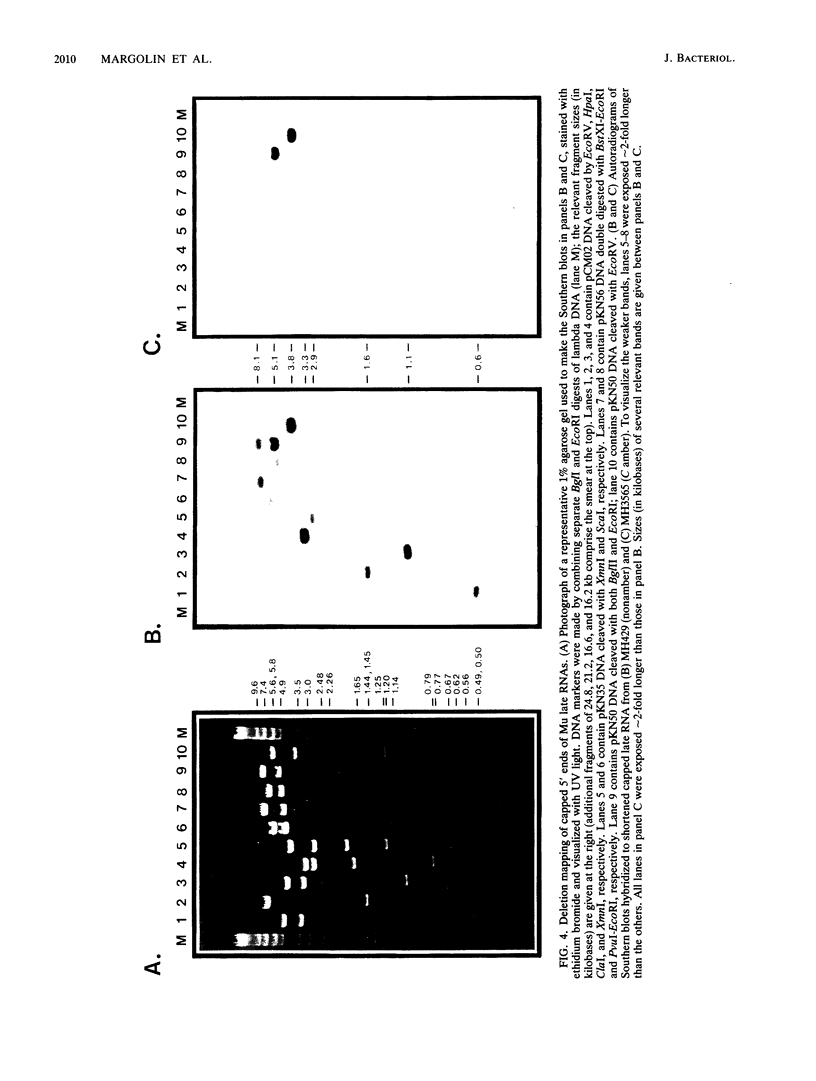
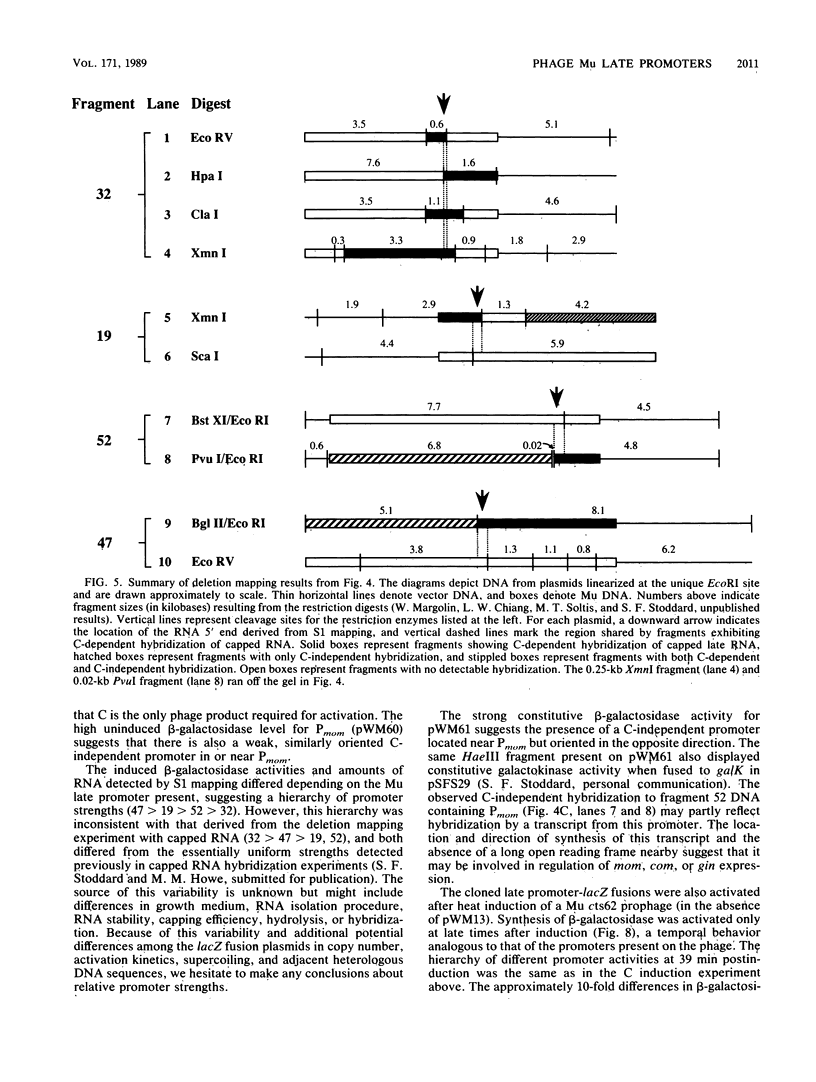
Late transcription of bacteriophage Mu, which results in the expression of phage morphogenetic functions, is dependent on Mu C protein. Earlier experiments indicated that Mu late RNAs originate from four promoters, including the previously characterized mom promoter. S1 nuclease protection experiments were used to map RNA 5' ends in the three new regions. Transcripts were initiated at these points only in the presence of C and were synthesized in a rightward direction on the Mu genome. Amber mutant marker rescue analysis of plasmid clones and limited DNA sequencing demonstrated that these new promoters are located between C and lys, upstream of I, and upstream of P within the N gene. A comparison of the promoter sequences upstream from the four RNA 5' ends yielded two conserved sequences: the first (tA . . cT, where capital and lowercase letters indicate 100 and 75% base conservation, respectively), at approximately -10, shares some similarity with the consensus Escherichia coli sigma 70 -10 region, while the second (ccATAAc CcCPuG/Cac, where Pu indicates a purine), in the -35 region, bears no resemblance to the E. coli -35 consensus. We propose that these conserved Mu late promoter consensus sequences are important for C-dependent promoter activity. Plasmids containing transcription fusions of these late promoters to lacZ exhibited C-dependent beta-galactosidase synthesis in vivo, and C was the only Mu product needed for this transactivation. As expected, the late promoter-lacZ fusions were activated only at late times after induction of a Mu prophage. The C-dependent activation of lacZ fusions containing only a few bases of the 5' end of Mu late RNA and the presence of altered promoter sequences imply that C acts at the level of transcription initiation.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhya S., Gottesman M., De Crombrugghe B. Release of polarity in Escherichia coli by gene N of phage lambda: termination and antitermination of transcription. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2534–2538. doi: 10.1073/pnas.71.6.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adley C. C., Bukhari A. I. Expression of the gin and mom genes of bacteriophage Mu. Gene. 1984 Apr;28(1):17–28. doi: 10.1016/0378-1119(84)90083-0. [DOI] [PubMed] [Google Scholar]
- Bade E. G. Asymmetric transcription of bacteriophage Mu-1. J Virol. 1972 Dec;10(6):1205–1207. doi: 10.1128/jvi.10.6.1205-1207.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedwell D. M., Nomura M. Feedback regulation of RNA polymerase subunit synthesis after the conditional overproduction of RNA polymerase in Escherichia coli. Mol Gen Genet. 1986 Jul;204(1):17–23. doi: 10.1007/BF00330181. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Brosius J., Ullrich A., Raker M. A., Gray A., Dull T. J., Gutell R. R., Noller H. F. Construction and fine mapping of recombinant plasmids containing the rrnB ribosomal RNA operon of E. coli. Plasmid. 1981 Jul;6(1):112–118. doi: 10.1016/0147-619x(81)90058-5. [DOI] [PubMed] [Google Scholar]
- Burton Z. F., Gross C. A., Watanabe K. K., Burgess R. R. The operon that encodes the sigma subunit of RNA polymerase also encodes ribosomal protein S21 and DNA primase in E. coli K12. Cell. 1983 Feb;32(2):335–349. doi: 10.1016/0092-8674(83)90453-1. [DOI] [PubMed] [Google Scholar]
- Bölker M., Wulczyn F. G., Kahmann R. Role of bacteriophage Mu C protein in activation of the mom gene promoter. J Bacteriol. 1989 Apr;171(4):2019–2027. doi: 10.1128/jb.171.4.2019-2027.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M., McGrath J., Waskell L. New RNA polymerase from Escherichia coli infected with bacteriophage T7. Nature. 1970 Oct 17;228(5268):227–231. doi: 10.1038/228227a0. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M., Pero J. Structure of a Bacillus subtilis bacteriophage SPO1 gene encoding RNA polymerase sigma factor. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1236–1240. doi: 10.1073/pnas.80.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dairi T., Inokuchi K., Mizuno T., Mizushima S. Positive control of transcription initiation in Escherichia coli. A base substitution at the Pribnow box renders ompF expression independent of a positive regulator. J Mol Biol. 1985 Jul 5;184(1):1–6. doi: 10.1016/0022-2836(85)90038-5. [DOI] [PubMed] [Google Scholar]
- DuBow M. S., Bukhari A. I. The proteins of bacteriophage mu: composition of the virion and biosynthesis in vivo during lytic growth. Prog Clin Biol Res. 1981;64:47–67. [PubMed] [Google Scholar]
- Falco S. C., Laan K. V., Rothman-Denes L. B. Virion-associated RNA polymerase required for bacteriophage N4 development. Proc Natl Acad Sci U S A. 1977 Feb;74(2):520–523. doi: 10.1073/pnas.74.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes D., Herskowitz I. Polarity suppression by the Q gene product of bacteriophage lambda. J Mol Biol. 1982 Oct 5;160(4):549–569. doi: 10.1016/0022-2836(82)90314-x. [DOI] [PubMed] [Google Scholar]
- Giphart-Gassler M., Plasterk R. H., van de Putte P. G inversion in bacteriophage Mu: a novel way of gene splicing. Nature. 1982 May 27;297(5864):339–342. doi: 10.1038/297339a0. [DOI] [PubMed] [Google Scholar]
- Gloor G., Chaconas G. Sequence of bacteriophage Mu N and P genes. Nucleic Acids Res. 1988 Jun 10;16(11):5211–5212. doi: 10.1093/nar/16.11.5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloor G., Chaconas G. The bacteriophage Mu N gene encodes the 64-kDa virion protein which is injected with, and circularizes, infecting Mu DNA. J Biol Chem. 1986 Dec 15;261(35):16682–16688. [PubMed] [Google Scholar]
- Grossman A. D., Erickson J. W., Gross C. A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984 Sep;38(2):383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Involvement of the invertible G segment in bacteriophage mu tail fiber biosynthesis. Virology. 1984 Apr 30;134(2):296–317. doi: 10.1016/0042-6822(84)90299-x. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Howe M. M. Morphogenetic structures present in lysates of amber mutants of bacteriophage Mu. Virology. 1985 Jun;143(2):485–504. doi: 10.1016/0042-6822(85)90388-5. [DOI] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., Ives J., Margolin W., Howe M. M. Regulation and expression of the bacteriophage mu mom gene: mapping of the transactivation (dad) function to the C region. Gene. 1985;39(1):71–76. doi: 10.1016/0378-1119(85)90109-x. [DOI] [PubMed] [Google Scholar]
- Hattman S., Ives J. S1 nuclease mapping of the phage Mu mom gene promoter: a model for the regulation of mom expression. Gene. 1984 Jul-Aug;29(1-2):185–198. doi: 10.1016/0378-1119(84)90179-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisig P., Kahmann R. The sequence and mom-transactivation function of the C gene of bacteriophage Mu. Gene. 1986;43(1-2):59–67. doi: 10.1016/0378-1119(86)90008-9. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Mahoney M. E., Wulff D. L., Rosenberg M. Identification of the DNA binding domain of the phage lambda cII transcriptional activator and the direct correlation of cII protein stability with its oligomeric forms. Genes Dev. 1988 Feb;2(2):184–195. doi: 10.1101/gad.2.2.184. [DOI] [PubMed] [Google Scholar]
- Ho Y. S., Wulff D. L., Rosenberg M. Bacteriophage lambda protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature. 1983 Aug 25;304(5928):703–708. doi: 10.1038/304703a0. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Howe M. M., O'Day K. J., Schultz D. W. Isolation of mutations defining five new cistrons essential for development of bacteriophage Mu. Virology. 1979 Mar;93(2):303–319. doi: 10.1016/0042-6822(79)90235-6. [DOI] [PubMed] [Google Scholar]
- Howe M. M. Prophage deletion mapping of bacteriophage Mu-1. Virology. 1973 Jul;54(1):93–101. doi: 10.1016/0042-6822(73)90118-9. [DOI] [PubMed] [Google Scholar]
- Irwin N., Ptashne M. Mutants of the catabolite activator protein of Escherichia coli that are specifically deficient in the gene-activation function. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8315–8319. doi: 10.1073/pnas.84.23.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahmann R., Seiler A., Wulczyn F. G., Pfaff E. The mom gene of bacteriophage mu: a unique regulatory scheme to control a lethal function. Gene. 1985;39(1):61–70. doi: 10.1016/0378-1119(85)90108-8. [DOI] [PubMed] [Google Scholar]
- Kamp D., Kahmann R., Zipser D., Broker T. R., Chow L. T. Inversion of the G DNA segment of phage Mu controls phage infectivity. Nature. 1978 Feb 9;271(5645):577–580. doi: 10.1038/271577a0. [DOI] [PubMed] [Google Scholar]
- Kassavetis G. A., Geiduschek E. P. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause H. M., Rothwell M. R., Higgins N. P. The early promoter of bacteriophage Mu: definition of the site of transcript initiation. Nucleic Acids Res. 1983 Aug 25;11(16):5483–5495. doi: 10.1093/nar/11.16.5483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwoh D. Y., Zipser D. Identification of the gin protein of bacteriophage mu. Virology. 1981 Oct 15;114(1):291–296. doi: 10.1016/0042-6822(81)90280-4. [DOI] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- Mahadevan S., Wright A. A bacterial gene involved in transcription antitermination: regulation at a rho-independent terminator in the bgl operon of E. coli. Cell. 1987 Jul 31;50(3):485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- Margolin W., Howe M. M. Localization and DNA sequence analysis of the C gene of bacteriophage Mu, the positive regulator of Mu late transcription. Nucleic Acids Res. 1986 Jun 25;14(12):4881–4897. doi: 10.1093/nar/14.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs C. F., Howe M. M. AvaII and BglI restriction maps of bacteriophage Mu. Virology. 1983 Apr 30;126(2):563–575. doi: 10.1016/s0042-6822(83)80013-0. [DOI] [PubMed] [Google Scholar]
- Martin S. A., Moss B. Modification of RNA by mRNA guanylyltransferase and mRNA (guanine-7-)methyltransferase from vaccinia virions. J Biol Chem. 1975 Dec 25;250(24):9330–9335. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mellado R. P., Barthelemy I., Salas M. In vivo transcription of bacteriophage phi 29 DNA early and late promoter sequences. J Mol Biol. 1986 Sep 20;191(2):191–197. doi: 10.1016/0022-2836(86)90256-1. [DOI] [PubMed] [Google Scholar]
- Meyer B. J., Ptashne M. Gene regulation at the right operator (OR) of bacteriophage lambda. III. lambda repressor directly activates gene transcription. J Mol Biol. 1980 May 15;139(2):195–205. doi: 10.1016/0022-2836(80)90304-6. [DOI] [PubMed] [Google Scholar]
- Miller K. W., Wu H. C. Cotranscription of the Escherichia coli isoleucyl-tRNA synthetase (ileS) and prolipoprotein signal peptidase (lsp) genes. Fine-structure mapping of the lsp internal promoter. J Biol Chem. 1987 Jan 5;262(1):389–393. [PubMed] [Google Scholar]
- Munson L. M., Stormo G. D., Niece R. L., Reznikoff W. S. lacZ translation initiation mutations. J Mol Biol. 1984 Aug 25;177(4):663–683. doi: 10.1016/0022-2836(84)90043-3. [DOI] [PubMed] [Google Scholar]
- O'Day K., Schultz D., Ericsen W., Rawluk L., Howe M. Correction and refinement of the genetic map of bacteriophage Mu. Virology. 1979 Mar;93(2):320–328. doi: 10.1016/0042-6822(79)90236-8. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Brinkman A., van de Putte P. DNA inversions in the chromosome of Escherichia coli and in bacteriophage Mu: relationship to other site-specific recombination systems. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5355–5358. doi: 10.1073/pnas.80.17.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R. H., Vollering M., Brinkman A., Van de Putte P. Analysis of the methylation-regulated Mu mom transcript. Cell. 1984 Jan;36(1):189–196. doi: 10.1016/0092-8674(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Plasterk R. H., Vrieling H., Van de Putte P. Transcription initiation of Mu mom depends on methylation of the promoter region and a phage-coded transactivator. Nature. 1983 Jan 27;301(5898):344–347. doi: 10.1038/301344a0. [DOI] [PubMed] [Google Scholar]
- Pramanik A., Wertheimer S. J., Schwartz J. J., Schwartz I. Expression of Escherichia coli infC: identification of a promoter in an upstream thrS coding sequence. J Bacteriol. 1986 Nov;168(2):746–751. doi: 10.1128/jb.168.2.746-751.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sanger F., Air G. M., Barrell B. G., Brown N. L., Coulson A. R., Fiddes C. A., Hutchison C. A., Slocombe P. M., Smith M. Nucleotide sequence of bacteriophage phi X174 DNA. Nature. 1977 Feb 24;265(5596):687–695. doi: 10.1038/265687a0. [DOI] [PubMed] [Google Scholar]
- Schumann W., Bade E. G., Forgie R. A., Howe M. M. Cloning of DNA fragments of the right end of phage mu and location of the HindIII, SalI, PstI, and BamHI restriction sites on the genetic map of mu. Virology. 1980 Jul 30;104(2):418–425. doi: 10.1016/0042-6822(80)90344-x. [DOI] [PubMed] [Google Scholar]
- Schumann W., Bade E. G. In vitro constructed plasmids containing both ends of bacteriophage Mu DNA express phage functions. Mol Gen Genet. 1979 Jan 16;169(1):97–105. doi: 10.1007/BF00267550. [DOI] [PubMed] [Google Scholar]
- Schumm J. W., Moore D. D., Blattner F. R., Howe M. M. Correlation of the genetic and physical maps in the central region of the bacteriophage Mu genome. Virology. 1980 Aug;105(1):185–195. doi: 10.1016/0042-6822(80)90166-x. [DOI] [PubMed] [Google Scholar]
- Shore S. H., Howe M. M. Bacteriophage Mu T mutants are defective in synthesis of the major head polypeptide. Virology. 1982 Jul 15;120(1):264–268. doi: 10.1016/0042-6822(82)90026-5. [DOI] [PubMed] [Google Scholar]
- Simatake H., Rosenberg M. Purified lambda regulatory protein cII positively activates promoters for lysogenic development. Nature. 1981 Jul 9;292(5819):128–132. doi: 10.1038/292128a0. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stoddard S. F., Howe M. M. DNA sequence within the Mu C operon. Nucleic Acids Res. 1987 Sep 11;15(17):7198–7198. doi: 10.1093/nar/15.17.7198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint A., Lecocq J. P. Sensitivity of bacteriophage Mu-1 development to rifampicin and streptolydigin. Mol Gen Genet. 1974 Mar 14;129(2):185–188. doi: 10.1007/BF00268631. [DOI] [PubMed] [Google Scholar]
- Van Leerdam E., Karreman C., van de Putte P. Ner, a cro-like function of bacteriophage Mu. Virology. 1982 Nov;123(1):19–28. doi: 10.1016/0042-6822(82)90291-4. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., Gassler M., Stevens W. F., van de Putte P. On the control of transcription of bacteriophage Mu. Mol Gen Genet. 1974;131(2):85–96. doi: 10.1007/BF00266145. [DOI] [PubMed] [Google Scholar]
- Wijffelman C., van de Putte P. Transcription of bacteriophage mu. An analysis of the transcription pattern in the early phase of phage development. Mol Gen Genet. 1974;135(4):327–337. doi: 10.1007/BF00271147. [DOI] [PubMed] [Google Scholar]
- de Boer H. A., Comstock L. J., Vasser M. The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc Natl Acad Sci U S A. 1983 Jan;80(1):21–25. doi: 10.1073/pnas.80.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]





