Abstract
5′-Adenylylsulfate (APS) reductase (EC 1.8.99.-) catalyzes the reduction of activated sulfate to sulfite in plants. The evidence presented here shows that a domain of the enzyme is a glutathione (GSH)-dependent reductase that functions similarly to the redox cofactor glutaredoxin. The APR1 cDNA encoding APS reductase from Arabidopsis thaliana is able to complement the cysteine auxotrophy of an Escherichia coli cysH [3′-phosphoadenosine-5′-phosphosulfate (PAPS) reductase] mutant, only if the E. coli strain produces glutathione. The purified recombinant enzyme (APR1p) can use GSH efficiently as a hydrogen donor in vitro, showing a Km[GSH] of ≈0.6 mM. Gene dissection was used to express separately the regions of APR1p from amino acids 73–327 (the R domain), homologous with microbial PAPS reductase, and from amino acids 328–465 (the C domain), homologous with thioredoxin. The R and C domains alone are inactive in APS reduction, but the activity is partially restored by mixing the two domains. The C domain shows a number of activities that are typical of E. coli glutaredoxin rather than thioredoxin. Both the C domain and APR1p are highly active in GSH-dependent reduction of hydroxyethyldisulfide, cystine, and dehydroascorbate, showing a Km[GSH] in these assays of ≈1 mM. The R domain does not show these activities. The C domain is active in GSH-dependent reduction of insulin disulfides and ribonucleotide reductase, whereas APR1p and R domain are inactive. The C domain can substitute for glutaredoxin in vivo as demonstrated by complementation of an E. coli mutant, underscoring the functional similarity between the two enzymes.
Plants and microorganisms are able to reduce sulfate to sulfide for synthesis of the thiol group of cysteine. Sulfate is first activated by ATP sulfurylase, forming 5′-adenylylsulfate (APS). APS can be phosphorylated by APS kinase, forming 3′-phosphoadenosine-5′-phosphosulfate (PAPS). Depending upon the organism, either APS or PAPS can be used for sulfate reduction. In general, it is thought that prokaryotes and fungi use PAPS, whereas photosynthetic eukaryotes use APS (1). However, the APS pathway is not universally accepted, and it has been argued that plants may reduce sulfate as do microorganisms by means of PAPS reductase (2). This enzyme, encoded by cysH in Escherichia coli, is dependent on the cofactors thioredoxin or glutaredoxin (3).
Key evidence supporting an APS-dependent pathway was obtained by cloning of three individual cDNAs (APR1, -2, and -3; accession nos. U43412, U56921, and U56922, respectively) for APS reductase (EC 1.8.99.-) from Arabidopsis thaliana (4, 5). This enzyme was termed a “reductase” because of its homology with the protein encoded by cysH; however, it is likely identical to the enzyme termed APS sulfotransferase, on the basis of their similar catalytic requirements and molecular weights (4, 6). The major difference is that the product of the sulfotransferase is believed to be an organic thiosulfate (1), whereas the reductase may produce sulfite (4), although the actual reaction product has not been determined for either enzyme. APS reductase is distinguished from the cysH product by its preference for APS over PAPS and by its ability to function in an E. coli thioredoxin/glutaredoxin double mutant (4). These properties may be due to its unusual two-domain structure, consisting of a reductase domain (R domain) at the amino terminus, showing homology with the cysH product; and a carboxyl-terminal region (C domain), showing homology with thioredoxin and glutaredoxin. This structure suggests that a dithiol–disulfide transhydrogenase mechanism may be involved in APS reduction. It should be noted that the APR-encoded enzyme shows no sequence homology with the APS reductase from dissimilatory sulfate-reducing bacteria, which is an iron–sulfur flavoenzyme (7). To avoid confusion we propose that the enzyme described in this report be referred to as the plant-type APS reductase.
Proteins belonging to the thioredoxin superfamily contain a reactive Cys pair that undergo reversible disulfide bond formation as electrons are transferred to a variety of reductases (3). Thioredoxin is specifically reduced by NADPH or ferredoxin-dependent thioredoxin reductase (8). Glutaredoxin, a related cofactor, is specifically reduced by glutathione (GSH), which is itself maintained in a reduced state by NADPH-dependent glutathione reductase (9). The specificity for GSH lies in a glutathione binding site in glutaredoxin that is absent from thioredoxin (10).
In this report, evidence is presented that GSH may be the electron source used by APS reductase. This result is significant because GSH was proposed to be the natural substrate of APS sulfotransferase in plants (11). Further, enzyme dissection revealed that the C domain of APS reductase shows GSH-dependent transhydrogenase activities that are characteristic of glutaredoxin. Finally, we report that the R and C domains, expressed as separate proteins, are not active in APS reduction, but the activity is reconstituted when they are mixed together. This result establishes the independent but interactive catalytic roles played by the APS reductase domains.
MATERIALS AND METHODS
Reagents and General Methods.
Nuclease P1 (catalog. no. N8630), Spirulina thioredoxin (T3658), insulin (I5500), and glutathione reductase (G4759) were from Sigma. Bovine thioredoxin reductase was purchased from IMCO (Stockholm). E. coli thioredoxin and thioredoxin reductase were the kind gift of Charles Williams (Veterans Affairs Medical Center, Ann Arbor, MI). E. coli ribonucleotide reductase was the kind gift of Joanne Stubbe (Massachusetts Institute of Technology, Cambridge). [35S]PAPS was purchased from New England Nuclear. [35S]APS was prepared by dephosphorylating [35S]PAPS (4).
Bacteriological media were prepared as described by Miller (12). Sambrook et al. (13) was followed for nucleic acid methods and Laemmli (14), for denaturing protein gel electrophoresis. Concentrations of pure proteins were measured from the difference in absorbance at 280 and 260 nm (15), and the concentrations of proteins in crude extracts were measured by the Bradford method (16).
Construction of Expression Plasmids and Purification of Recombinant Proteins.
For protein purification the Novagen S-Tag system was used. The APR1-encoded protein (APR1p) expression plasmid (pET-APR1) was prepared by cloning a 1420-bp EcoRI–SalI fragment from the APR1 cDNA into pET-30a. The C-domain expression plasmid (pET-C) was prepared by cloning the 380-bp EcoRV–SalI fragment of APR1 into pET-30a. A plasmid was prepared from which C-domain expression is regulated by an arabinose-inducible promoter (pBAD-C) by cloning the ≈400-bp XbaI–HindIII fragment from pET-C into pBAD33 (17). The R-domain expression plasmid (pET-R) was prepared by removing the 380-bp EcoRI–SalI C-domain fragment from pET-APR1. pET-R carries a 1040-bp R-domain fragment and produces a protein that includes up to amino acid 348 in APR1p. The recombinant C domain includes amino acids 349 to 465 of APR1p. The APR2 expression plasmid was prepared by cloning a 1450-bp EcoRI fragment from APR2 into pET-30a. The APR3 expression plasmid was prepared by cloning a 1500-bp EcoRI fragment from APR3 into pET-30b.
BL21(DE3)plysS was transformed with the pET expression plasmids. Transformants were grown at 37°C to an optical density (at 600 nm) of 0.6–0.8 and expression was induced with 1 mM isopropyl β-d-thiogalactoside (IPTG) for 3 h. The cells from 100-ml culture were resuspended in 10 ml of 150 mM NaCl/0.1% (vol/vol) Triton X-100/20 mM Tris⋅HCl, pH 7.5, and were then lysed by a freeze–thaw cycle followed by sonication. The lysate was centrifuged (14,000 × g for 10 min) and the supernatant was filtered through a 0.4-μm-pore membrane. The lysate was incubated with 2 ml of S-protein agarose for 30 min. The agarose was washed with 5 vol of lysis buffer and the bound protein was released by overnight digestion with enterokinase, which was subsequently removed by using Ekapture (Novagen) agarose. The supernatant containing the purified recombinant protein was dialyzed against 100 mM Tris⋅HCl, pH 8.5, and stored at −70°C. On average ≈800 μg of recombinant protein was recovered.
Glutaredoxin was purified from E. coli DHB4 transformed with pBAD-grxA (grxA cloned into pBAD18; ref. 17). The purification was carried out as in ref. 18, except that ultrafiltration replaced the gel filtration step. No contaminating proteins were visible on an overloaded SDS/PAGE gel.
Strain Construction and Complementation Assays.
The E. coli strains used in this study are described in Table 1. TL1, -2, and -3 were prepared by transduction (12) of JM96 with phage P1 lysates prepared in JF518, JTG10, and A304, respectively. Transduction was carried out with infective P1kc prepared by growing KL39 at 42°C. M9 medium with leucine, tryptophan, histidine, arginine, and thiamin, with or without cysteine, was used for complementation analysis of cysH strains with pλYES-APR1 (4). The inocula for the experiment in Fig. 1 were transferred once on cysteine-free medium. Glutathione, γ-glutamylcysteine (γ-EC), and cysteine were measured by HPLC using fluorescent labeling with monobromobimane (23). Complementation of FA47 and FA87 was carried out with pBAD-C or pBAD33-grxA as a positive control. These strains are derived from E. coli strain DHB4. FA87 carries a complementing TrxC plasmid containing the counterselectable wild-type rpsL allele. After transformation with either pBAD-C or pBAD33-grxA the transformants were counterselected on medium containing streptomycin sulfate (selection for colonies that have lost the TrxC plasmid). The colonies were then tested for arabinose-dependent complementation on medium containing 0.2% (wt/vol) arabinose. Rich medium was used for FA87 and minimal medium lacking cysteine was used for FA47. Arabinose-dependent expression of the C domain was confirmed by immunoblotting with an S-Tag antibody.
Table 1.
E. coli strains
| Strain | Relevant genotype | Origin |
|---|---|---|
| JM96 | cysH56 | CGSC* |
| KL39 | P1kc+ | CGSC* |
| JF518 | gshB∷kan | (19) |
| JTG10 | gshA∷Tn-miniKan | (20) |
| A304 | trxB15∷kan | (21) |
| TL1 | cysH56, gshB∷km | This study |
| TL2 | cysH56, gshA∷km | This study |
| TL3 | cysH56, trxB15∷km | This study |
| DHB4 | Δ(ara–leu)7697 | (22) |
| FA87 | ΔtrxA, trxC, grxA∷km | † |
| pBAD39-trxC | ||
| Δ(ara–leu)7697 | ||
| FA47 | ΔtrxA, grxA∷km | † |
| Δ(ara714–leu)∷Tn10 |
Coli Genetic Stock Center (Yale University).
E. J. Stewart, F.Å, and J. Beckwith, unpublished work.
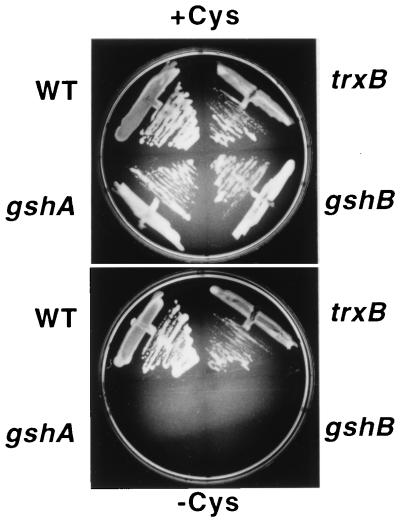
Figure 1.
Complementation of E. coli cysH by plant APS reductase requires glutathione. WT refers to the cysH strain carrying wild-type alleles for gshA, gshB, and trxB. The others are cysH strains carrying the designated mutation. The relevant medium nutrient composition is indicated above and below the photographs. The cultures were incubated for 48 h at 30°C. Thiol compounds and APS reductase activity were measured in extracts prepared from the cells grown on +cysteine medium. The concentrations of thiol compounds are given below in nmol per g of fresh weight and APS reductase activity is given in pmol⋅min−1 per mg of protein. WT: GSH = 383, γ-EC not detected, cysteine not detected, and APS reductase = 215. trxB: GSH = 575, γ-EC not detected, cysteine not detected, and APS reductase = 4950. gshA: GSH not detected, γ-EC not detected, cysteine = 10, and APS reductase = 210. gshB: GSH = 6, γ-EC = 70, cysteine = 13, and APS reductase = 225
Enzyme Assays.
Unless indicated otherwise, the unit of enzyme activity is defined as μmol of product formed per min. Apparent Km values were calculated by least-squares nonlinear regression analysis (24). All values were determined from at least three independent experiments.
APS reductase was measured at 30°C (4). The 100-μl reaction mixture contained 100 mM Tris⋅HCl (pH 8.5), 500 mM Na2SO4, 1 mM EDTA, 25 μM [35S]APS (≈500 Bq⋅nmol−1), pure APS reductase or protein extract, and a hydrogen donor (DTT, GSH, cysteine, γ-EC, or 1 mM NADPH and 6 units of thioredoxin reductase). The reaction was started by addition of the hydrogen donor. The amount of sulfur dioxide formed in a reaction lacking the hydrogen donor (the background rate) was subtracted from the complete reaction mix to obtain the catalytic rate. To avoid large changes in substrate concentration the incubation time and protein concentration were adjusted so that the amount of product formed was at least 5-fold above the background but no more than 10% of the substrate in the reaction. In some cases the reaction was allowed to progress to completion to confirm that the enzyme can convert all the substrate to product.
GSH-dependent reduction of hydroxyethyldisulfide (HED) or cystine was measured in a 500-μl reaction mixture containing 0.8 mM HED or cystine, 0.35 mM NADPH, 2 units of glutathione reductase, 1.5 mM EDTA, 100 mM Tris⋅HCl (pH 8.0), and GSH (25). APRp was added, and the absorbance at 340 nm was measured continuously for 2 min at 24°C.
GSH-dependent reduction of dehydroascorbate (DHA) was measured in a 500-μl reaction mixture containing 2.0 mM DHA, 0.2 mM NADPH, 2 units of glutathione reductase, 0.27 mM EDTA, 50 mM sodium phosphate (pH 6.9), and GSH (26). The mixture, lacking DHA and APRp, was preincubated for 5 min. DHA and then APRp were added, and the absorbance at 340 nm was measured continuously for 2 min at 24°C.
In the HED, cystine, and DHA assays the rate of NADPH oxidation in the absence of APRp served as the background rate. All the reductase assays were tested with 6 units of thioredoxin reductase replacing GSH and glutathione reductase, but no activity was observed with this alternate reducing system.
The reduction of insulin disulfides (27) was measured in a 600-μl reaction mixture containing 2 mM EDTA, 100 mM sodium phosphate (pH 6.5), 0.13 mM insulin, 20 μM thioredoxin, GrxA or APRp, and a hydrogen source (1 mM DTT; 1 mM NADPH and 6 units of thioredoxin reductase; or 5 mM GSH, 1 mM NADPH, and 6 units of glutathione reductase). The increase in absorbance at 650 nm was monitored continuously at 24°C.
The reduction of ribonucleotide reductase disulfides was determined by measuring its activity (28). The 400-μl reaction mixture contained 30 mM Hepes (pH 7.6), 10 mM MgCl2, 1.5 mM EDTA, 0.5 mg⋅ml−1 BSA, 1.3 mM ATP, 0.05 unit of ribonucleotide reductase, 1 mM NADPH, cofactor (thioredoxin, GrxA, or APRp), and a reducing system (6 units of thioredoxin reductase or 5 mM GSH and 6 units of glutathione reductase). Ribonucleotide reductase was prereduced by incubation for 60 min on ice with 5 mM DTT. The DTT was removed by gel filtration on Sephadex G-25. The reaction was started by addition of 1 mM CDP. The decrease in absorbance at 340 nm was monitored continuously at 24°C.
RESULTS
Determination of the Hydrogen Donor for APS Reductase.
The APR cDNAs encoding APS reductase are able to complement the cysteine auxotrophy of an E. coli cysH mutant strain (4). The structure of APS reductase raised the question of what reducing system is used during complementation of cysH. The availability of glutathione and thioredoxin reductase mutants of E. coli allowed us to explore this question. Mutant alleles for gshA (γ-glutamylcysteine synthetase), gshB (glutathione synthetase), and trxB (thioredoxin reductase) were separately introduced into the cysH strain, and the derivatives were used to test the ability of APR1 to complement cysH. Fig. 1 shows that even though there is APS reductase activity in all the strains APR1 is able to complement cysH only if the strain contains glutathione.
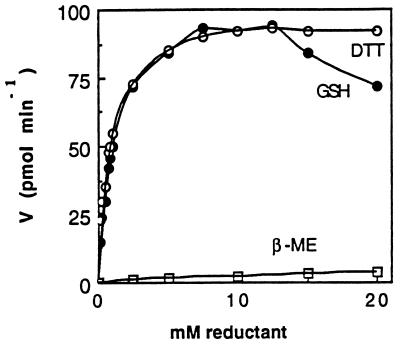
Pure recombinant APS reductases (APR1p, APR2p, and APR3p) were used to study the hydrogen donor requirement in vitro. The APR1p preparation is shown in Fig. 2, lane 2. APR2p and APR3p showed a similar size and level of purity (not shown). Fig. 3 shows the APS reductase activity of APR1p with three different electron donors. GSH is as effective as DTT in promoting APS reduction, whereas 2-mercaptoethanol is ineffective. At high concentrations GSH is slightly inhibitory. The GSH titration shows that Vmax of APR1p is 0.18 μmol⋅min−1⋅mg−1 achieved with 8–10 mM GSH. Similar results were obtained with the other APS reductases, although APR2p shows a significantly greater Vmax (Table 2). Irrespective of their specific activity, all the APR proteins show a similar Km value for GSH ranging from 0.6 to 1.2 mM. Cysteine and γ-EC were also tested as hydrogen sources for the APR proteins (data not shown). Cysteine did not function as a hydrogen donor, but the Vmax achieved with γ-EC was comparable to that with GSH, although the Km[γ-EC] was 9-fold greater than Km[GSH]. NADPH in combination with E. coli or bovine thioredoxin reductase and with or without E. coli or Spirulina thioredoxin was unable to promote APS reductase activity in any of the APR enzymes. A variety of conditions were tested. In particular, it was found that under the APS reductase assay conditions (pH 8.5, 500 mM Na2SO4) the NADPH/thioredoxin/thioredoxin reductase system retains ≈80% of its activity as measured by using the insulin disulfide reduction assay, so APS reduction would have been observed if the APR enzymes were able to use this system as a hydrogen source.
Figure 2.

Purified recombinant APR1p, R domain, and C domain. Lane 1, molecular mass standards; lane 2, 2 μg of APR1p; lane 3, 2 μg of R domain; lane 4, molecular mass standards; and lane 5, 1 μg of C domain. Lanes 1–3 and 4–5 were run on 10% and 12.5% polyacrylamide gels, respectively. The mass in kDa of the recombinant proteins predicted from the amino acid sequence and based on gel mobility (in parentheses) is as follows: APR1p = 53.7 (51 and 49); R domain = 40.1 (41 and 39.5); and C domain = 13.7 (12.8). The migration of APR1p and R domain as a doublet is likely due to inefficient cleavage with enterokinase. The molecular masses of standards in kDa are 200, 97.4, 68, 43, 29, 18.4, and 14.3.
Figure 3.
Hydrogen donor requirement of APS reductase. The activity from 0.60 μg of APR1p was measured with various amounts of DTT (○), GSH (•) or 2-mercaptoethanol (□). The assays were incubated for 20 min. Each point is the mean of three assays. Similar results were obtained with four different APR1p preparations.
Table 2.
Reductase assays and kinetic constants
| Assay | Protein | Km[GSH], mM | Vmax, μmol⋅min−1⋅mg−1 |
|---|---|---|---|
| APS | APR1p | 0.6 | 0.18 |
| APR2p | 1.2* | 3.10 | |
| APR3p | 0.9* | 0.21 | |
| HED | APR1p | 1.2 | 21.2 |
| C | 1.2 | 60.5 | |
| R | NA | NA | |
| Cystine | APR1p | 1.2 | 41.1 |
| C | 1.1* | 54.3 | |
| R | NA | NA | |
| DHA | APR1p | 1.1* | 52.8 |
| C | 1.2* | 62.4 | |
| R | NA | NA |
Assay refers to the substrate reduced; Km[GSH] was measured at saturating level of the second substrate indicated in the assay column. Vmax is in μmol⋅min−1 per mg of protein; NA, no activity or unable to be calculated. All values were calculated from four independent data sets. ∗ indicates SD 10–20% of value; all others less than 10% of value.
Analysis of the Function of APS Reductase Domains.
The function of the APS reductase domains was studied by expressing and purifying each as a separate recombinant polypeptide. The preparations used in the experiments are shown in Fig. 2, lanes 3 and 5. The isolated R (reductase) and C (carboxyl-terminal) domains were found to have low APS reductase activity, but this activity was not catalytic, because it was not proportional to the amount of protein added to the reaction (Fig. 4). DTT alone or in combination with thioredoxin or glutaredoxin did not promote APS reductase activity for either domain. When the R and C domains were combined, APS reductase activity was reconstituted, and this activity showed the features that are expected of a reaction catalyzed by two enzymes (Fig. 4). At a fixed level of R domain the reaction rate is directly proportional to the amount of C domain in the reaction up to a point beyond which the rate did not increase further. At the highest levels of C domain the R domain is limiting, the reaction rate being proportional to the amount of R domain in the reaction. A specific activity for the R domain of approximately 0.022–0.025 μmol⋅min−1⋅mg−1 can be calculated from the data. This is 1/6 to 1/8 the Vmax of the intact APR1p enzyme. The molar ratio of R and C domains needed to achieve maximal activity is approximately 1:60 compared with a 1:1 ratio for the intact enzyme. It is tempting to speculate that the high molar ratio and decreased catalytic efficiency is because of inefficient interaction of the separated R and C domains, but it is also possible that expression as recombinant protein fragments has resulted in preparations that are only partially active. However, the results show that the APS reductase domains play independent and interactive roles in catalysis.
Figure 4.
APS reductase activity of isolated APR1p domains. □, R domain; ▪, C domain. Various amounts of C domain were added to a reaction with 0.5 μl (○) or 1 μl (•) of R domain. The reaction mixtures contained 10 mM GSH. The concentration of the protein solutions was 20 pmol⋅μl−1.
Glutaredoxin Function of the C Domain of APS Reductase.
The homology of the C domain with disulfide-active cofactors, coupled with the finding that APS reductase uses GSH as a source of protons, suggests that the C domain may function as glutaredoxin, a GSH-disulfide oxidoreductase (3, 9). Glutaredoxin has several characteristic activities, including the ability to catalyze GSH-dependent reduction of small molecules such as HED, cystine, and DHA (24, 25). The C domain was found to be very active in HED, cystine, and DHA reduction (Table 2), with activities that are comparable to the activity of glutaredoxin. APR1p is also active in these reactions, but the R domain is inactive. When GSH and glutathione reductase are replaced with thioredoxin reductase the C domain is unable to catalyze the reduction of the small molecules. The Km[GSH] of the C domain and APR1p in these assays is ≈1 mM, similar to the Km[GSH] of the APR enzymes in the APS reductase assay. In total, the results suggest that it is the C domain of APS reductase that mediates the interaction of the enzyme with GSH by functioning as a glutaredoxin.
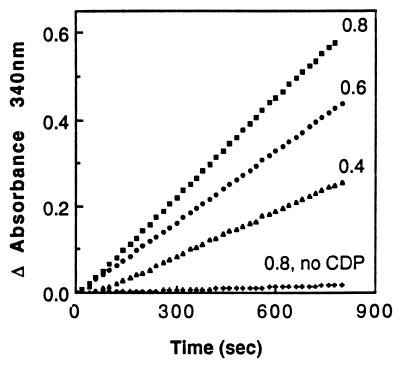
Both thioredoxin and glutaredoxin have the ability to reduce the disulfides of insulin (27). In the presence of DTT as a hydrogen source the C domain was found to be able to reduce insulin, whereas the R domain and APR1p were found to be inactive (Fig. 5). A similar level of activity for the C domain and GrxA could be obtained when DTT was replaced with GSH, NADPH, and glutathione reductase, whereas thioredoxin was inactive with this reducing system (not shown). Another activity associated with thioredoxin and glutaredoxin is the ability to serve as a hydrogen donor for ribonucleotide reductase (27). The C domain, in combination with GSH, NADPH, and glutathione reductase, is able to serve as a hydrogen donor for ribonucleotide reductase (Fig. 6). The reaction is dependent on the presence of CDP, the ribonucleotide reductase substrate, and the rate is proportional to the amount of C-domain added. This result indicates that under these conditions (up to 0.8 mM C domain and excess ribonucleotide reductase) the activity of the C domain limits the reaction rate. Table 3 shows that with GSH, glutathione reductase, and NADPH as the reducing system the C domain and glutaredoxin are able to promote ribonucleotide reductase activity but thioredoxin is unable to do so. The specific activity of the C domain is ≈1/7 that of GrxA. With thioredoxin reductase and NADPH as the hydrogen source only thioredoxin is able to promote ribonucleotide reductase activity. Taken together, these results demonstrate that the C domain is able to catalyze GSH-dependent reduction of protein disulfides in a fashion similar to that of glutaredoxin.
Figure 5.
The C domain can reduce the disulfides of insulin. The activity was measured with 20 μM of the enzyme indicated on the graph. The thioredoxin is from E. coli. The activity of the R domain and APR1p was not greater than the spontaneous rate in a reaction lacking protein. The assay measures the precipitation of the insulin β chain, detected as an increase in turbidity at 650 nm. A reaction with 20 μM glutaredoxin (Grx1) gave a curve that lies between the curves of thioredoxin and C domain (not shown).
Figure 6.
C domain can serve as the hydrogen donor for ribonucleotide reductase. The rate of NADPH oxidation is presented as the change in absorbance at 340 nm. The assay was carried out with three different concentrations of C domain, indicated on the graph in μM. The background rate was a reaction with 0.8 μM C domain but lacking the ribonucleotide reductase substrate CDP.
Table 3.
Ribonucleotide reductase activity with C domain, glutaredoxin, or thioredoxin
| Protein | Reducing system | Activity, nmol/10 min |
|---|---|---|
| C domain | GSH/GR | 24.4 |
| GrxA | GSH/GR | 172.4 |
| Thioredoxin | GSH/GR | NA |
| C-domain | TR | NA |
| GrxA | TR | NA |
| Thioredoxin | TR | 93.8 |
C domain, GrxA, and E. coli thioredoxin were 0.8 μM. GSH/GR refers to glutathione reductase and GSH, and TR refers to the thioredoxin reductase system. Activity is in nmol of NADP+ per 10 min. NA, no activity.
As with the insulin reduction assay, APR1p and the R domain are inactive with ribonucleotide reductase. This result is in contrast to the results with small molecules, where APR1p showed activity comparable to that of the isolated C domain. Thus it appears that reduction of protein disulfides by the C domain is blocked when it is physically associated with the R domain.
The ability of the C-domain to serve as a protein disulfide reductase was tested by a complementation assay using an E. coli strain carrying mutations in trxA, trxC, and grxA. Strain FA87 is absolutely dependent on a complementing plasmid encoding a functional disulfide reductase (E. J. Stewart, F.Å., and J. Beckwith, unpublished work). For example, it can grow if it carries a GrxA-expression plasmid but is unable to grow with GrxC, another E. coli glutaredoxin, which is a poor disulfide reductant compared with GrxA (29). Thus, FA87 offers a convenient way of assessing the in vivo disulfide reducing capacity of a test protein. The C-domain plasmid was able to complement strain FA87 in an arabinose-dependent manner, demonstrating in vivo the disulfide reductase activity of the C domain. However, the growth rate was slower than with the GrxA plasmid. This difference could reflect the lower efficiency of the C domain to serve as a reductant of ribonucleotide reductase (Table 3). We also tested whether the C-domain plasmid can complement another in vivo redox function attributed to thioredoxin and glutaredoxin, the ability to serve as a hydrogen donor for PAPS reductase (strain FA47). The C-domain plasmid was unable to complement this strain, indicating that it is unable to drive PAPS reductase activity. This result may reflect a difference between the C domain and GrxA with respect to specificity and/or reducing properties.
DISCUSSION
Evidence is presented that APS reductase uses GSH as a hydrogen donor and that its C domain shows properties that resemble those of glutaredoxin. The evidence also shows that the catalytic mechanism of APS reductase depends on the interaction of two domains with distinct functions.
The simplest explanation for the ability of APR1 to complement the cysH mutation only if the strain produces glutathione is that APS reductase uses GSH as a hydrogen donor. The key finding that bolsters this hypothesis is that the C domain of APS reductase functions like glutaredoxin, a well characterized GSH-dependent oxidoreductase.
Glutaredoxin serves as a cofactor for a number of reductases, including ribonucleotide reductase (3); thus, knowledge of its function could be invaluable in proposing a catalytic mechanism for APS reductase. There are two proposed mechanisms for glutaredoxin. The first, for reduction of protein disulfides, requires the two active site Cys residues, and an intramolecular disulfide with the substrate protein is formed as an intermediate (18). In the case of ribonucleotide reductase, glutaredoxin reduces a disulfide, which charges the reductase with the protons needed for ribonucleotide reduction (30). In this model, it is possible that the C domain may function in transferring protons to the R domain, which then directly catalyzes APS reduction. The R domain of all the APR proteins contain two Cys pairs that could be reduced by the C domain (CEPC, amino acids 294–297; and CC, amino acids 202–203 in APR1p). However, the mechanism is unlikely to be a simple transfer of protons, because E. coli thioredoxin or glutaredoxin was unable to substitute for the C domain in promoting APS reduction by the R domain. This finding suggests that there is specificity in the interaction between the R and C domains.
It is intriguing that APR1p is unable to catalyze insulin precipitation or serve as a hydrogen donor for ribonucleotide reductase, whereas it is able to reduce small disulfide molecules. This could be due to steric inhibition of the C domain by the R domain, or it might reflect a fundamental difference in catalytic mechanism between protein disulfide reduction and the reduction of small molecules. Indeed, reduction of small molecule disulfides by glutaredoxin is thought to proceed by a mechanism different from reduction of protein disulfides. This mechanism requires only one of the two active site Cys residues, the reactive Cys (Cys-14 in T4 Grx) and involves the formation of an intermediate mixed disulfide with glutathione (18). The high activity of APS reductase in reducing small molecules raises the interesting possibility that the C domain could function in reducing S-sulfoglutathione generated by the R domain or S-sulfo derivatives of the R domain. Another intriguing possibility is that APS reductase could play a role outside sulfate assimilation, in reducing a variety of small disulfide molecules or S-sulfo compounds generated spontaneously when sulfite, a toxic and reactive compound, accumulates in plastids, such as during sulfur dioxide exposure (31).
Thioredoxin and glutaredoxin share similar active sites and have similar secondary structures (3). However, it is the GSH binding site of glutaredoxins that is a key structural feature of this class of cofactors (10, 32). Alignment of the C-domain sequence with the glutaredoxins reported in ref. 10 indicates that it contains many of the amino acids that constitute the conserved GSH binding site. Moreover, the predicted secondary structure of the C domain closely matches that of the glutaredoxins. This match suggests that the C domain may contain a GSH binding site.
Glutathione is a major intracellular thiol compound of plant cells, so it could serve as the natural hydrogen donor for APS reductase. The glutathione concentration in plastids, where APS reductase is localized, has been variously reported between 3 and 10 mM, and this compound is maintained predominantly in the reduced state (33, 34). Thus the apparent Km[GSH] of APS reductase reported here and in reference 6 (≈1 mM) is below the physiological concentration. γ-EC is a minor thiol compound in plants, so it could not serve as a proton source for APS reductase. Significantly, GSH was reported to be the most likely substrate for APS sulfotransferase (11). The experiments reported here do not specifically rule out the possibility that another factor such as chloroplast thioredoxin reductase could serve to reduce APS reductase. However, this study and that reported in ref. 6 have established that APS reductase, in the presence of physiological concentrations of GSH, can function without such accessory factors. This finding is in agreement with the early work showing that ATP and GSH are the only required factors for the reduction of sulfate to sulfite in plant chloroplasts (35).
The finding that the separated APS reductase domains can catalyze APS reduction prompts the idea that this enzyme could be post-translationally processed into two proteins. We do not think this is the case because antibodies against purified APR1p react with a protein from A. thaliana leaves of ≈48 kDa on immunoblots (not shown). This is close to the expected molecular mass of the protein after transport into plastids.
With the cloning of APS reductase from higher plants (4) and the work reported here, long-standing questions about the nature of sulfate reduction in plants have been resolved. First, the data confirm the early work on an APS-dependent enzyme that is able to function with only reduced thiol compounds as a hydrogen donor (36). Although recently purified APS sulfotransferase (6) has not yet been directly confirmed to be identical to APS reductase, the similarities of these enzymes make this highly likely. The present work also provides a mechanistic basis for the ability of the enzyme to use GSH directly for APS reduction, namely that APS reductase carries its own cofactor domain that functions as a glutaredoxin.
Acknowledgments
We thank Drs. Bruce Demple, James Fuchs, Peter Model, and Marjorie Russel for providing bacterial strains and Drs. JoAnne Stubbe and Charles Williams for providing purified proteins. This work was funded by National Science Foundation Grant IBN-9601145. F.Å. is supported by a fellowship from the European Molecular Biology Organization.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: GSH, reduced glutathione (when the reduced form is not specifically being referred to, the term “glutathione” is used); γ-EC, γ-glutamylcysteine; APS, 5′-adenylylsulfate; PAPS, 3′-phosphoadenosine-5′-phosphosulfate; DHA, dehydroascorbate; HED, hydroxyethyldisulfide.
References
- 1.Schiff J A. In: Encyclopedia of Plant Physiology. Läuchli A, Bieleski R L, editors. 15A. Heidelberg: Springer; 1983. pp. 401–421. [Google Scholar]
- 2.Schiffmann S, Schwenn J D. FEBS Lett. 1994;355:229–232. doi: 10.1016/0014-5793(94)01193-1. [DOI] [PubMed] [Google Scholar]
- 3.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 4.Setya A, Murillo M, Leustek T. Proc Natl Acad Sci USA. 1996;93:13383–13388. doi: 10.1073/pnas.93.23.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gutierrez-Marcos J, Roberts M A, Campbell E I, Wray J L. Proc Natl Acad Sci USA. 1996;93:13377–13382. doi: 10.1073/pnas.93.23.13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanno N, Nagahisa E, Sato M, Sato Y. Planta. 1996;198:440–446. [Google Scholar]
- 7.Speich N, Dahl C, Heisig P, Klein A, Lottspeich F, Stetter K O, Trüper H G. Microbiology (Reading, UK) 1994;140:1273–1284. doi: 10.1099/00221287-140-6-1273. [DOI] [PubMed] [Google Scholar]
- 8.Buchanan B B, Schürmann P, Decottignies P, Lozano R M. Arch Biochem Biophys. 1994;314:257–260. doi: 10.1006/abbi.1994.1439. [DOI] [PubMed] [Google Scholar]
- 9.Fuchs J A. In: Glutathione—Chemical, Biochemical and Medical Aspects, Coenzymes and Cofactors. Dolphin D, Poulson R, Avramovic O, editors. 3, Part B. New York: Wiley; 1989. pp. 551–570. [Google Scholar]
- 10.Nikkola M, Gleason F K, Saarinen M, Joelson T, Bjornberg O, Eklund H. J Biol Chem. 1991;266:16105–16112. [PubMed] [Google Scholar]
- 11.Tsang M L-S, Schiff J A. Plant Sci Lett. 1978;11:177–183. [Google Scholar]
- 12.Miller J H. Experiments in Molecular Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1972. [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 14.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 15.Segel I H. Biochemical Calculations. 2nd Ed. New York: Wiley; 1976. , Appendix II. [Google Scholar]
- 16.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 17.Guzman L-M, Belin D, Carson M J, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bushweller J H, Åslund F, Wüthrich K, Holmgren A. Biochemistry. 1992;31:9288–9293. doi: 10.1021/bi00153a023. [DOI] [PubMed] [Google Scholar]
- 19.Daws T, Lim C-J, Fuchs J. J Bacteriol. 1989;171:5218–5221. doi: 10.1128/jb.171.9.5218-5221.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg J T, Demple B. J Bacteriol. 1986;168:1026–1029. doi: 10.1128/jb.168.2.1026-1029.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Russel M, Model P. In: Thioredoxin and Glutaredoxin Systems: Structure and Function. Holmgren A, Brändén C-I, Jörnvall H, Sjöberg B-M, editors. New York: Raven; 1986. pp. 331–337. [Google Scholar]
- 22.Boyd D, Manoil C, Beckwith J. Proc Natl Acad Sci USA. 1987;84:8525–8529. doi: 10.1073/pnas.84.23.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fahey R C, Newton G L. Methods Enzymol. 1987;143:85–96. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- 24.Brooks S P J. BioTechniques. 1992;13:906–911. [PubMed] [Google Scholar]
- 25.Holmgren A. J Biol Chem. 1979;254:3664–3671. [PubMed] [Google Scholar]
- 26.Trümper S, Follmann H, Häberlein I. FEBS Lett. 1994;352:159–162. doi: 10.1016/0014-5793(94)00947-3. [DOI] [PubMed] [Google Scholar]
- 27.Holmgren A. J Biol Chem. 1979;254:9267–9632. [Google Scholar]
- 28.Holmgren A. J Biol Chem. 1979;254:3672–3678. [PubMed] [Google Scholar]
- 29.Åslund F, Nordstrand K, Berndt K D, Nikkola M, Bergman T, Ponstingl H, Jörnvall H, Otting G, Holmgren A. J Biol Chem. 1996;271:6736–6745. doi: 10.1074/jbc.271.12.6736. [DOI] [PubMed] [Google Scholar]
- 30.Mao S-S, Holler T P, Yu G X, Bollinger J M, Jr, Booker S, Johnston M I, Stubbe J. Biochemistry. 1992;31:9733–9743. doi: 10.1021/bi00155a029. [DOI] [PubMed] [Google Scholar]
- 31.Würfel M, Häberlein I, Follmann H. FEBS Lett. 1990;268:146–148. doi: 10.1016/0014-5793(90)80994-t. [DOI] [PubMed] [Google Scholar]
- 32.Bushweller J H, Billeter M, Holmgren A, Wüthrich K. J Mol Biol. 1994;235:1585–1597. doi: 10.1006/jmbi.1994.1108. [DOI] [PubMed] [Google Scholar]
- 33.Anderson J W, Foyer C H, Walker D A. Biochim Biophys Acta. 1983;724:69–74. [Google Scholar]
- 34.Foyer C H, Halliwell B. Planta. 1976;133:21–25. doi: 10.1007/BF00386001. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt A, Trebst A. Biochim Biophys Acta. 1969;180:529–535. doi: 10.1016/0005-2728(69)90031-0. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt A. Arch Microbiol. 1972;84:77–86. doi: 10.1007/BF00408084. [DOI] [PubMed] [Google Scholar]