Abstract
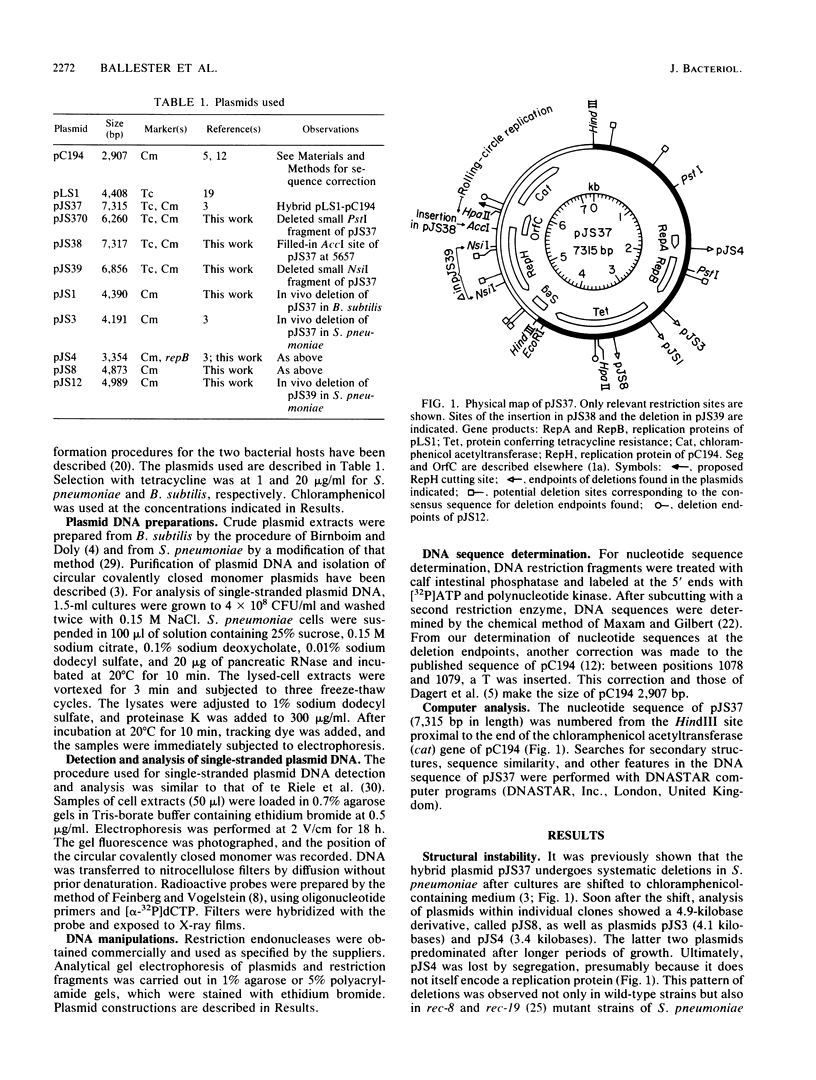
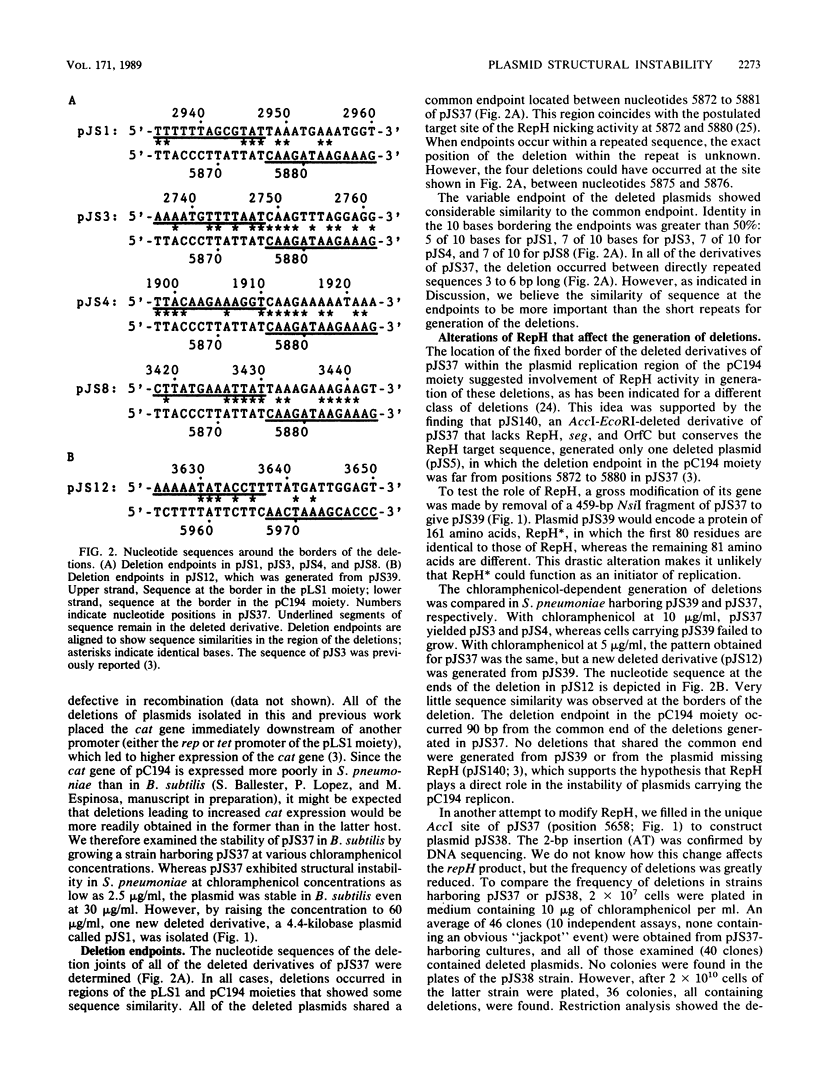
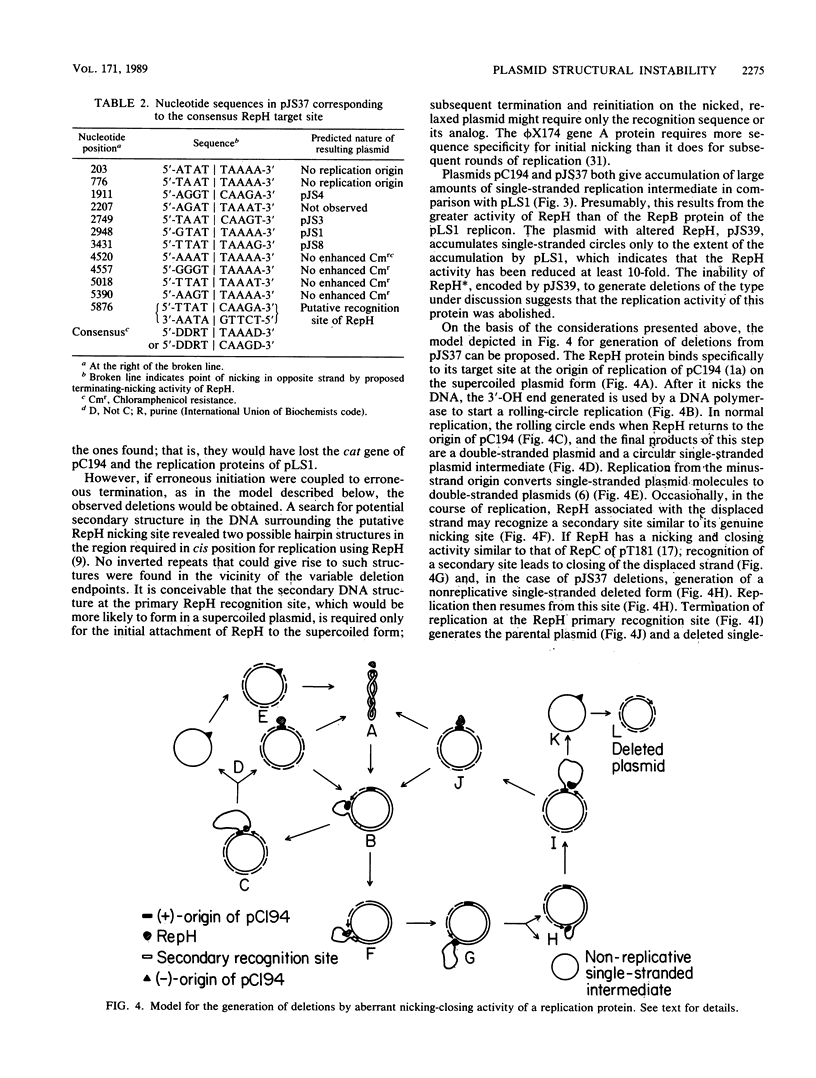
The hybrid plasmid pJS37 is composed of the streptococcal plasmid pLS1, which confers tetracycline resistance, and the staphylococcal plasmid pC194, which confers chloramphenicol resistance. When gram-positive bacteria containing pJS37 were grown in the presence of chloramphenicol, four different deleted derivatives accumulated. The deletions in the plasmid enhanced resistance to chloramphenicol by placing the cat gene of pC194 near promoters of pLS1. All four deletions shared a common endpoint that corresponded to the putative target site for DNA strand nicking by the pC194 replication protein, RepH. At the other, variable endpoint, the DNA sequence was similar to the putative RepH target sequence. Alteration of the RepH protein, by in vitro modification of the gene encoding it, eliminated this class of deletions. By extending a previously proposed model for the generation of a different but related class of deletions (B. Michel and S.D. Ehrlich, EMBO J. 5:3691-3696, 1986), a comprehensive model that could generate both classes of deletions is suggested. It proposes that a nicking-closing activity of the plasmid replication protein at its normal target site and, aberrantly, at sites with similar sequence can generate deletions either proximal or distal to the aberrant site during rolling-circle replication of the plasmid.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Alonso J. C., Tailor R. H. Initiation of plasmid pC194 replication and its control in Bacillus subtilis. Mol Gen Genet. 1987 Dec;210(3):476–484. doi: 10.1007/BF00327200. [DOI] [PubMed] [Google Scholar]
- Baas P. D. DNA replication of single-stranded Escherichia coli DNA phages. Biochim Biophys Acta. 1985 Jun 24;825(2):111–139. doi: 10.1016/0167-4781(85)90096-x. [DOI] [PubMed] [Google Scholar]
- Ballester S., Lopez P., Alonso J. C., Espinosa M., Lacks S. A. Selective advantage of deletions enhancing chloramphenicol acetyltransferase gene expression in Streptococcus pneumoniae plasmids. Gene. 1986;41(2-3):153–163. doi: 10.1016/0378-1119(86)90094-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Jones I., Goze A., Romac S., Niaudet B., Ehrlich S. D. Replication functions of pC194 are necessary for efficient plasmid transduction by M13 phage. EMBO J. 1984 Jan;3(1):81–86. doi: 10.1002/j.1460-2075.1984.tb01764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa M., Lopez P., Perez-Ureña M. T., Lacks S. A. Interspecific plasmid transfer between Streptococcus pneumoniae and Bacillus subtilis. Mol Gen Genet. 1982;188(2):195–201. doi: 10.1007/BF00332675. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gros M. F., te Riele H., Ehrlich S. D. Rolling circle replication of single-stranded DNA plasmid pC194. EMBO J. 1987 Dec 1;6(12):3863–3869. doi: 10.1002/j.1460-2075.1987.tb02724.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A. D., Ross H. F., Novick R. P. Functional analysis of a palindromic sequence required for normal replication of several staphylococcal plasmids. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2165–2169. doi: 10.1073/pnas.84.8.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Dubnau D. Analysis of plasmid deletional instability in Bacillus subtilis. J Bacteriol. 1985 Jun;162(3):1014–1023. doi: 10.1128/jb.162.3.1014-1023.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Aoki K., Naito A. Illegitimate recombination mediated in vitro by DNA gyrase of Escherichia coli: structure of recombinant DNA molecules. Proc Natl Acad Sci U S A. 1982 Jun;79(12):3724–3728. doi: 10.1073/pnas.79.12.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iordanescu S., Surdeanu M. Isolation and complementation of temperature-sensitive replication mutants of Staphylococcus aureus plasmid pC194. Mol Gen Genet. 1983;191(2):201–206. doi: 10.1007/BF00334814. [DOI] [PubMed] [Google Scholar]
- Jones I. M., Primrose S. B., Ehrlich S. D. Recombination between short direct repeats in a recA host. Mol Gen Genet. 1982;188(3):486–489. doi: 10.1007/BF00330053. [DOI] [PubMed] [Google Scholar]
- Koepsel R. R., Khan S. A. Cleavage of single-stranded DNA by plasmid pT181-encoded RepC protein. Nucleic Acids Res. 1987 May 26;15(10):4085–4097. doi: 10.1093/nar/15.10.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koepsel R. R., Murray R. W., Rosenblum W. D., Khan S. A. The replication initiator protein of plasmid pT181 has sequence-specific endonuclease and topoisomerase-like activities. Proc Natl Acad Sci U S A. 1985 Oct;82(20):6845–6849. doi: 10.1073/pnas.82.20.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacks S. A., Lopez P., Greenberg B., Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986 Dec 20;192(4):753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966 Jan;53(1):207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P., Espinosa M., Greenberg B., Lacks S. A. Generation of deletions in pneumococcal mal genes cloned in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5189–5193. doi: 10.1073/pnas.81.16.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciag I. E., Viret J. F., Alonso J. C. Replication and incompatibility properties of plasmid pUB110 in Bacillus subtilis. Mol Gen Genet. 1988 May;212(2):232–240. doi: 10.1007/BF00334690. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination at the replication origin of bacteriophage M13. Proc Natl Acad Sci U S A. 1986 May;83(10):3386–3390. doi: 10.1073/pnas.83.10.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B., Ehrlich S. D. Illegitimate recombination occurs between the replication origin of the plasmid pC194 and a progressing replication fork. EMBO J. 1986 Dec 20;5(13):3691–3696. doi: 10.1002/j.1460-2075.1986.tb04701.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Adler G. K., Projan S. J., Carleton S., Highlander S. K., Gruss A., Khan S. A., Iordanescu S. Control of pT181 replication I. The pT181 copy control function acts by inhibiting the synthesis of a replication protein. EMBO J. 1984 Oct;3(10):2399–2405. doi: 10.1002/j.1460-2075.1984.tb02146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyet A., del Solar G. H., Espinosa M. Identification of the origin and direction of replication of the broad-host-range plasmid pLS1. Nucleic Acids Res. 1988 Jan 11;16(1):115–133. doi: 10.1093/nar/16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth M. J., Brown D. R., Hurwitz J. Analysis of bacteriophage phi X174 gene A protein-mediated termination and reinitiation of phi X DNA synthesis. II. Structural characterization of the covalent phi X A protein-DNA complex. J Biol Chem. 1984 Aug 25;259(16):10556–10568. [PubMed] [Google Scholar]
- Stassi D. L., Lopez P., Espinosa M., Lacks S. A. Cloning of chromosomal genes in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7028–7032. doi: 10.1073/pnas.78.11.7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Solar G. H., Puyet A., Espinosa M. Initiation signals for the conversion of single stranded to double stranded DNA forms in the streptococcal plasmid pLS1. Nucleic Acids Res. 1987 Jul 24;15(14):5561–5580. doi: 10.1093/nar/15.14.5561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- te Riele H., Michel B., Ehrlich S. D. Single-stranded plasmid DNA in Bacillus subtilis and Staphylococcus aureus. Proc Natl Acad Sci U S A. 1986 Apr;83(8):2541–2545. doi: 10.1073/pnas.83.8.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mansfeld A. D., van Teeffelen H. A., Fluit A. C., Baas P. D., Jansz H. S. Effect of SSB protein on cleavage of single-stranded DNA by phi X gene A protein and A* protein. Nucleic Acids Res. 1986 Feb 25;14(4):1845–1861. doi: 10.1093/nar/14.4.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]