Abstract
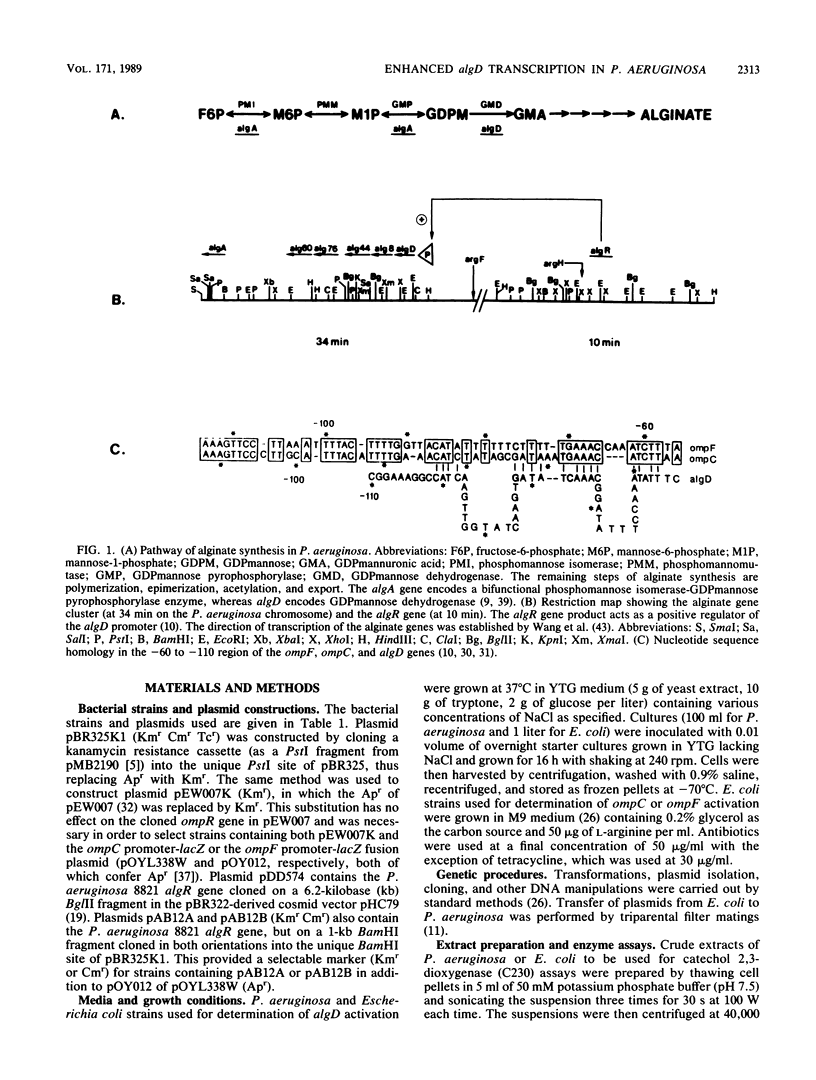
Chronic lung infection with mucoid, alginate-producing strains of Pseudomonas aeruginosa is a major cause of mortality in cystic fibrosis (CF) patients. Transcriptional activation of the P. aeruginosa algD gene, which encodes GDPmannose dehydrogenase, is essential for alginate synthesis. Activation of algD is dependent on the product of the algR gene. Sequence homology between the P. aeruginosa algR gene and the Escherichia coli ompR gene, which regulates the cellular response to changes in osmolarity of the growth medium, together with the abnormally high levels of Na+ and Cl- in respiratory tract fluid in CF patients suggested that high osmolarity in the lung of the CF patient might be a signal contributing to the induction of alginate synthesis (mucoidy) in infecting P. aeruginosa. In both mucoid and nonmucoid P. aeruginosa strains (containing a functional algR gene), transcriptional activation of algD increased as the osmolarity of the culture medium increased. The increased activation of algD at high osmolarity was not in itself sufficient to induce alginate synthesis in nonmucoid strains, however, suggesting that other environmental factors are involved in full activation of the alginate genes. The targets of AlgR and OmpR, the algD promoter and the ompC and ompF promoters, respectively, were found to have appreciable sequence homology in the -60 to -110 regions. In E. coli, OmpR was capable of activating the algD promoter nearly as well as AlgR, but in both cases, activation occurred only under conditions of high osmolarity.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Cairney J., Booth I. R., Higgins C. F. Osmoregulation of gene expression in Salmonella typhimurium: proU encodes an osmotically induced betaine transport system. J Bacteriol. 1985 Dec;164(3):1224–1232. doi: 10.1128/jb.164.3.1224-1232.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Darzins A., Chakrabarty A. M. Cloning of genes controlling alginate biosynthesis from a mucoid cystic fibrosis isolate of Pseudomonas aeruginosa. J Bacteriol. 1984 Jul;159(1):9–18. doi: 10.1128/jb.159.1.9-18.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzins A., Frantz B., Vanags R. I., Chakrabarty A. M. Nucleotide sequence analysis of the phosphomannose isomerase gene (pmi) of Pseudomonas aeruginosa and comparison with the corresponding Escherichia coli gene manA. Gene. 1986;42(3):293–302. doi: 10.1016/0378-1119(86)90233-7. [DOI] [PubMed] [Google Scholar]
- Darzins A., Wang S. K., Vanags R. I., Chakrabarty A. M. Clustering of mutations affecting alginic acid biosynthesis in mucoid Pseudomonas aeruginosa. J Bacteriol. 1985 Nov;164(2):516–524. doi: 10.1128/jb.164.2.516-524.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Dikshit R., Konyecsni W. M., Chakrabarty A. M., Misra T. K. The algR gene, which regulates mucoidy in Pseudomonas aeruginosa, belongs to a class of environmentally responsive genes. J Bacteriol. 1989 Mar;171(3):1278–1283. doi: 10.1128/jb.171.3.1278-1283.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Gene algD coding for GDPmannose dehydrogenase is transcriptionally activated in mucoid Pseudomonas aeruginosa. J Bacteriol. 1987 Jan;169(1):351–358. doi: 10.1128/jb.169.1.351-358.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Gill J. F., Chakrabarty A. M. Pseudomonas aeruginosa infection in cystic fibrosis: nucleotide sequence and transcriptional regulation of the algD gene. Nucleic Acids Res. 1987 Jun 11;15(11):4567–4581. doi: 10.1093/nar/15.11.4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V., Tomasek P., Darzins A., Chakrabarty A. M. Gene amplification induces mucoid phenotype in rec-2 Pseudomonas aeruginosa exposed to kanamycin. J Bacteriol. 1986 Feb;165(2):510–516. doi: 10.1128/jb.165.2.510-516.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doggett R. G. Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl Microbiol. 1969 Nov;18(5):936–937. doi: 10.1128/am.18.5.936-937.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill J. F., Deretic V., Chakrabarty A. M. Overproduction and assay of Pseudomonas aeruginosa phosphomannose isomerase. J Bacteriol. 1986 Aug;167(2):611–615. doi: 10.1128/jb.167.2.611-615.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govan J. R., Harris G. S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986 Oct;3(10):302–308. [PubMed] [Google Scholar]
- Hall M. N., Silhavy T. J. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol. 1981 Sep 5;151(1):1–15. doi: 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- Higgins C. F., Dorman C. J., Stirling D. A., Waddell L., Booth I. R., May G., Bremer E. A physiological role for DNA supercoiling in the osmotic regulation of gene expression in S. typhimurium and E. coli. Cell. 1988 Feb 26;52(4):569–584. doi: 10.1016/0092-8674(88)90470-9. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Holloway B. W. Genetics of Pseudomonas. Bacteriol Rev. 1969 Sep;33(3):419–443. doi: 10.1128/br.33.3.419-443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IACOCCA V. F., SIBINGA M., BARBERO G. J. RESPIRATORY TRACT BACTERIOLOGY IN CYSTIC FIBROSIS. Am J Dis Child. 1963 Sep;106:315–324. doi: 10.1001/archpedi.1963.02080050317012. [DOI] [PubMed] [Google Scholar]
- Jo Y. L., Nara F., Ichihara S., Mizuno T., Mizushima S. Purification and characterization of the OmpR protein, a positive regulator involved in osmoregulatory expression of the ompF and ompC genes in Escherichia coli. J Biol Chem. 1986 Nov 15;261(32):15252–15256. [PubMed] [Google Scholar]
- Jovanovich S. B., Martinell M., Record M. T., Jr, Burgess R. R. Rapid response to osmotic upshift by osmoregulated genes in Escherichia coli and Salmonella typhimurium. J Bacteriol. 1988 Feb;170(2):534–539. doi: 10.1128/jb.170.2.534-539.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilbourn J. P. Bacterial content and ionic composition of sputum in cystic fibrosis. Lancet. 1978 Feb 11;1(8059):334–334. doi: 10.1016/s0140-6736(78)90111-3. [DOI] [PubMed] [Google Scholar]
- McPherson M. A., Dormer R. L. The molecular and biochemical basis of cystic fibrosis. Biosci Rep. 1987 Mar;7(3):167–185. doi: 10.1007/BF01124787. [DOI] [PubMed] [Google Scholar]
- McPherson M. A., Goodchild M. C. The biochemical defect in cystic fibrosis. Clin Sci (Lond) 1988 Apr;74(4):337–345. doi: 10.1042/cs0740337. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Chou M. Y., Inouye M. A comparative study on the genes for three porins of the Escherichia coli outer membrane. DNA sequence of the osmoregulated ompC gene. J Biol Chem. 1983 Jun 10;258(11):6932–6940. [PubMed] [Google Scholar]
- Mizuno T., Mizushima S. Characterization by deletion and localized mutagenesis in vitro of the promoter region of the Escherichia coli ompC gene and importance of the upstream DNA domain in positive regulation by the OmpR protein. J Bacteriol. 1986 Oct;168(1):86–95. doi: 10.1128/jb.168.1.86-95.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T., Wurtzel E. T., Inouye M. Cloning of the regulatory genes (ompR and envZ) for the matrix proteins of the Escherichia coli outer membrane. J Bacteriol. 1982 Jun;150(3):1462–1466. doi: 10.1128/jb.150.3.1462-1466.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa A. J., Ninfa E. G., Lupas A. N., Stock A., Magasanik B., Stock J. Crosstalk between bacterial chemotaxis signal transduction proteins and regulators of transcription of the Ntr regulon: evidence that nitrogen assimilation and chemotaxis are controlled by a common phosphotransfer mechanism. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5492–5496. doi: 10.1073/pnas.85.15.5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norioka S., Ramakrishnan G., Ikenaka K., Inouye M. Interaction of a transcriptional activator, OmpR, with reciprocally osmoregulated genes, ompF and ompC, of Escherichia coli. J Biol Chem. 1986 Dec 25;261(36):17113–17119. [PubMed] [Google Scholar]
- O'Hoy K., Krishnapillai V. Recalibration of the Pseudomonas aeruginosa strain PAO chromosome map in time units using high-frequency-of-recombination donors. Genetics. 1987 Apr;115(4):611–618. doi: 10.1093/genetics/115.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa Y., Mizuno T., Mizushima S. Roles of the Pribnow box in positive regulation of the ompC and ompF genes in Escherichia coli. J Bacteriol. 1987 Mar;169(3):1331–1334. doi: 10.1128/jb.169.3.1331-1334.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds H. Y., Di Sant'Agnese P. A., Zierdt C. H. Mucoid Pseudomonas aeruginosa. A sign of cystic fibrosis in young adults with chronic pulmonary disease? JAMA. 1976 Nov 8;236(19):2190–2192. doi: 10.1001/jama.236.19.2190. [DOI] [PubMed] [Google Scholar]
- Slauch J. M., Garrett S., Jackson D. E., Silhavy T. J. EnvZ functions through OmpR to control porin gene expression in Escherichia coli K-12. J Bacteriol. 1988 Jan;170(1):439–441. doi: 10.1128/jb.170.1.439-441.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sá-Correia I., Darzins A., Wang S. K., Berry A., Chakrabarty A. M. Alginate biosynthetic enzymes in mucoid and nonmucoid Pseudomonas aeruginosa: overproduction of phosphomannose isomerase, phosphomannomutase, and GDP-mannose pyrophosphorylase by overexpression of the phosphomannose isomerase (pmi) gene. J Bacteriol. 1987 Jul;169(7):3224–3231. doi: 10.1128/jb.169.7.3224-3231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wang S. K., Sa'-Correia I., Darzins A., Chakrabarty A. M. Characterization of the Pseudomonas aeruginosa alginate (alg) gene region II. J Gen Microbiol. 1987 Aug;133(8):2303–2314. doi: 10.1099/00221287-133-8-2303. [DOI] [PubMed] [Google Scholar]


