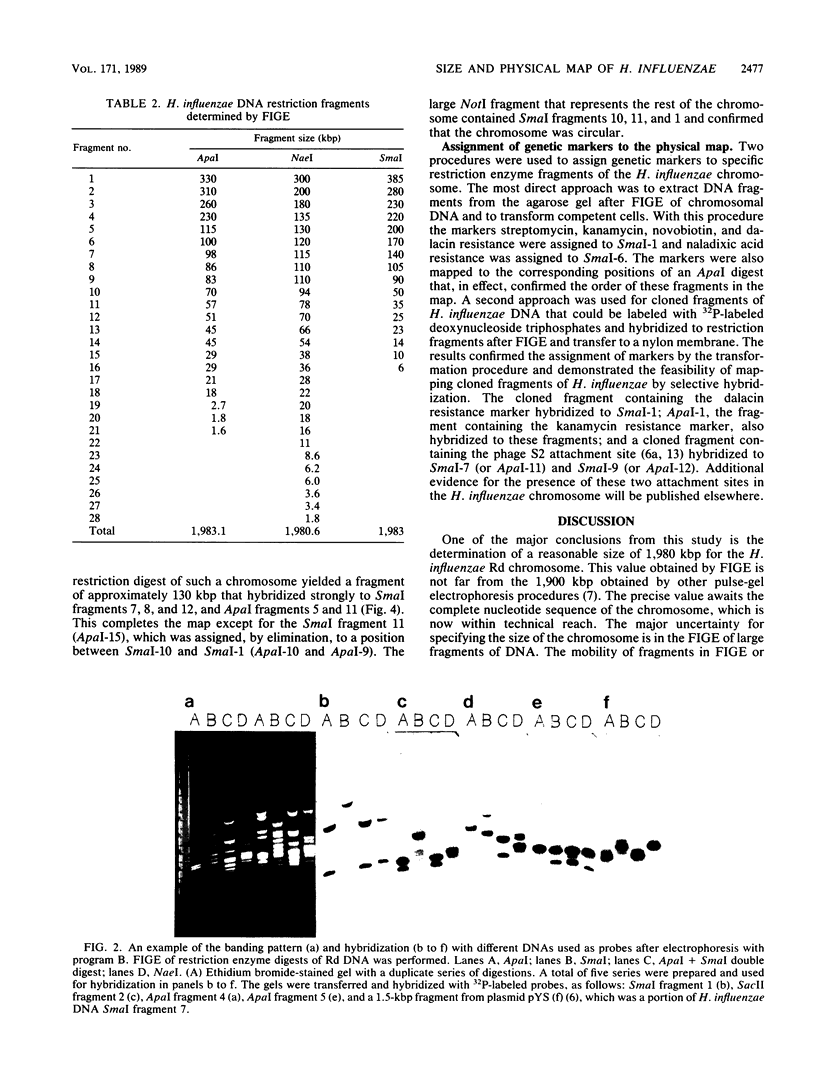
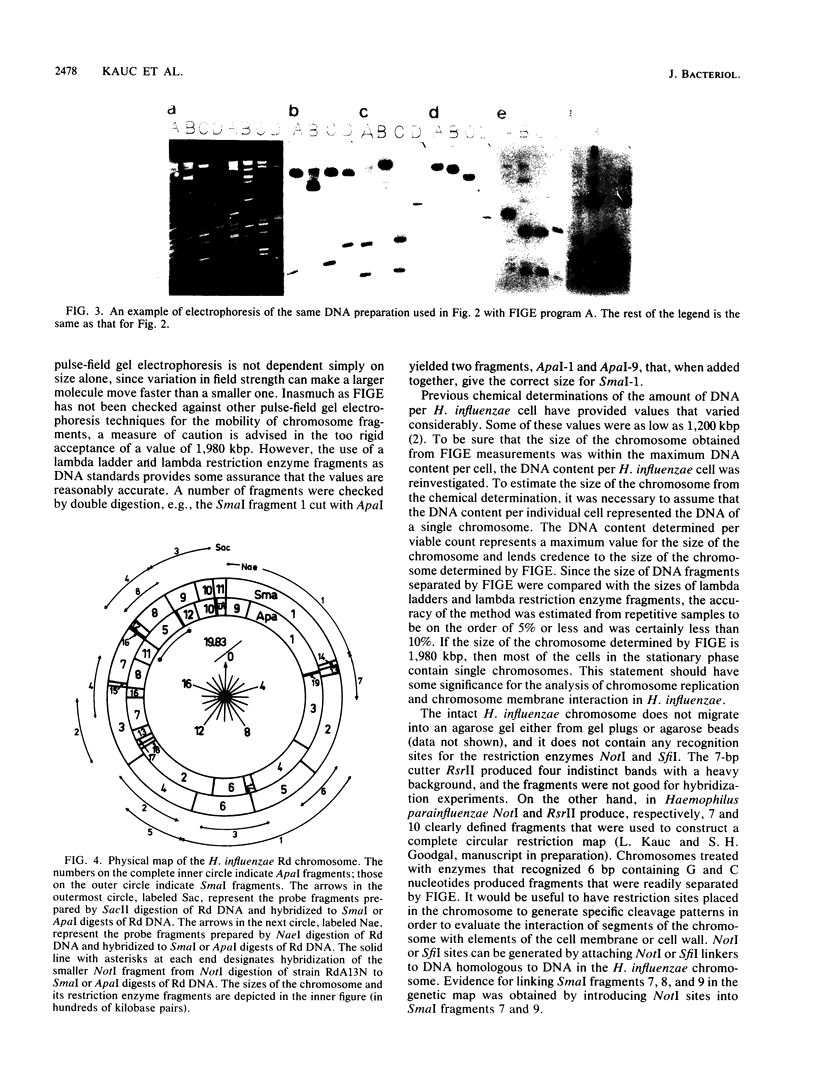
Abstract
A variation of pulse-field electrophoresis, field-inversion gel electrophoresis, was used to determine the size and physical map of the chromosome of Haemophilus influenzae. The DNA of H. influenzae had a low G + C content (39%) and no restriction sites for the enzymes NotI or SfiI. However, a number of restriction enzymes (SmaI, ApaI, NaeI, and SacII) that recognized 6-base-pair sequences containing only G and C nucleotides were found to generate a reasonable number of DNA fragments that were separable in agarose gels by field-inversion gel electrophoresis. The sizes of the DNA fragments were calibrated with a lambda DNA ladder and lambda DNA restriction fragments. The sum of fragment sizes obtained with restriction digests yielded a value for the chromosome of 1,980 kilobase pairs. Hybridization of a labeled fragment with two or more fragments from a digest with a different restriction enzyme provided the information needed to construct a circular map of the H. influenzae chromosome.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALEXANDER H. E., LEIDY G. Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951 Apr 1;93(4):345–359. doi: 10.1084/jem.93.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERNS K. I., THOMAS C. A., Jr ISOLATION OF HIGH MOLECULAR WEIGHT DNA FROM HEMOPHILUS INFLUENZAE. J Mol Biol. 1965 Mar;11:476–490. doi: 10.1016/s0022-2836(65)80004-3. [DOI] [PubMed] [Google Scholar]
- Cantor C. R., Smith C. L., Mathew M. K. Pulsed-field gel electrophoresis of very large DNA molecules. Annu Rev Biophys Biophys Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Jackson D. A., Cook P. R. A general method for preparing chromatin containing intact DNA. EMBO J. 1985 Apr;4(4):913–918. doi: 10.1002/j.1460-2075.1985.tb03718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauc L., Goodgal S. H. Amplification of DNA at a prophage attachment site in Haemophilus influenzae. J Bacteriol. 1989 Apr;171(4):1898–1903. doi: 10.1128/jb.171.4.1898-1903.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O. Sizing of the Haemophilus influenzae Rd genome by pulsed-field agarose gel electrophoresis. J Bacteriol. 1988 Sep;170(9):4402–4405. doi: 10.1128/jb.170.9.4402-4405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalka J., Goodgal S. H. Genetic and physical map of the chromosome of Hemophilus influenzae. J Mol Biol. 1969 Oct 28;45(2):407–421. doi: 10.1016/0022-2836(69)90115-6. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Stuy J. H. Transfer of genetic information within a colony of Haemophilus influenzae. J Bacteriol. 1985 Apr;162(1):1–4. doi: 10.1128/jb.162.1.1-4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Fitzmaurice W. P., Scocca J. J. Integration of the bacteriophage HP1c1 genome into the Haemophilus influenzae Rd chromosome in the lysogenic state. J Bacteriol. 1986 Jan;165(1):297–300. doi: 10.1128/jb.165.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury P. G., Lane M. J. Generation of lambda phage concatemers for use as pulsed field electrophoresis size markers. Nucleic Acids Res. 1987 May 11;15(9):3930–3930. doi: 10.1093/nar/15.9.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZAMENHOF S., ALEXANDER H. E., LEIDY G. Studies on the chemistry of the transforming activity. I. Resistance to physical and chemical agents. J Exp Med. 1953 Oct;98(4):373–397. doi: 10.1084/jem.98.4.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoon K. C., Scocca J. J. Constitution of the cell envelope of Haemophilus influenzae in relation to competence for genetic transformation. J Bacteriol. 1975 Aug;123(2):666–677. doi: 10.1128/jb.123.2.666-677.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]





