Abstract
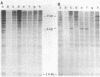
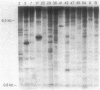
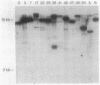
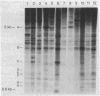
Extensive restriction-fragment-length polymorphism was revealed in Escherichia coli strains by using a region of the bacteriophage M13 genome as a DNA hybridization probe. This variation was observed across natural strains, in clinical samples, and to a lesser extent in laboratory strains. The sequence in M13 which revealed this fingerprint pattern was a region of the gene III coat protein, which contains two clusters of a 15-base-pair repeat. Oligonucleotides made to a consensus of these repeats also revealed the fingerprint profile. While this consensus sequence has significant homology to the lambda chi site sequence, an oligonucleotide made of the chi sequence did not reveal polymorphic fingerprint patterns in E. coli. The strain variation revealed by the M13 and M13-derived oligonucleotide probes will be useful for bacterial characterization and should find use in studies of bacterial evolution and population dynamics. The findings raise questions about what these repeated sequences are and why they are so variable.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Devor E. J., Ivanovich A. K., Hickok J. M., Todd R. D. A rapid method for confirming cell line identity: DNA "fingerprinting" with a minisatellite probe from M13 bacteriophage. Biotechniques. 1988 Mar;6(3):200–202. [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Kelly R., Taylor B. A., Bulfield G. Mouse DNA 'fingerprints': analysis of chromosome localization and germ-line stability of hypervariable loci in recombinant inbred strains. Nucleic Acids Res. 1987 Apr 10;15(7):2823–2836. doi: 10.1093/nar/15.7.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Wilson V., Thein S. L. Hypervariable 'minisatellite' regions in human DNA. Nature. 1985 Mar 7;314(6006):67–73. doi: 10.1038/314067a0. [DOI] [PubMed] [Google Scholar]
- Lewin R. DNA fingerprints in health and disease. Science. 1986 Aug 1;233(4763):521–522. doi: 10.1126/science.3726544. [DOI] [PubMed] [Google Scholar]
- Malone R. E., Chattoraj D. K., Faulds D. H., Stahl M. M., Stahl F. W. Hotspots for generalized recombination in the Escherichia coli chromosome. J Mol Biol. 1978 Jun 5;121(4):473–491. doi: 10.1016/0022-2836(78)90395-9. [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Leppert M., O'Connell P., Wolff R., Holm T., Culver M., Martin C., Fujimoto E., Hoff M., Kumlin E. Variable number of tandem repeat (VNTR) markers for human gene mapping. Science. 1987 Mar 27;235(4796):1616–1622. doi: 10.1126/science.3029872. [DOI] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Evidence for clonal population structure in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jan;81(1):198–201. doi: 10.1073/pnas.81.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Selander R. K. Standard reference strains of Escherichia coli from natural populations. J Bacteriol. 1984 Feb;157(2):690–693. doi: 10.1128/jb.157.2.690-693.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryskov A. P., Jincharadze A. G., Prosnyak M. I., Ivanov P. L., Limborska S. A. M13 phage DNA as a universal marker for DNA fingerprinting of animals, plants and microorganisms. FEBS Lett. 1988 Jun 20;233(2):388–392. doi: 10.1016/0014-5793(88)80467-8. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stern M. J., Ames G. F., Smith N. H., Robinson E. C., Higgins C. F. Repetitive extragenic palindromic sequences: a major component of the bacterial genome. Cell. 1984 Jul;37(3):1015–1026. doi: 10.1016/0092-8674(84)90436-7. [DOI] [PubMed] [Google Scholar]
- Vassart G., Georges M., Monsieur R., Brocas H., Lequarre A. S., Christophe D. A sequence in M13 phage detects hypervariable minisatellites in human and animal DNA. Science. 1987 Feb 6;235(4789):683–684. doi: 10.1126/science.2880398. [DOI] [PubMed] [Google Scholar]
- Wong Z., Wilson V., Jeffreys A. J., Thein S. L. Cloning a selected fragment from a human DNA 'fingerprint': isolation of an extremely polymorphic minisatellite. Nucleic Acids Res. 1986 Jun 11;14(11):4605–4616. doi: 10.1093/nar/14.11.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]