Abstract
Helicobacter pylori is one of the world's most successful human pathogens causing gastric ulcers and cancers. A key virulence factor of H. pylori is the Cag pathogenicity island, which encodes a type IV secretion system. HP0525 is an essential component of the Cag system and acts as an inner membrane associated ATPase. HP0525 forms double hexameric ring structures, with the C-terminal domains (CTDs) forming a closed ring and the N-terminal domains (NTDs) forming a dynamic, open ring. Here, the crystal structure of HP0525 in complex with a fragment of HP1451, a protein of previously unknown function, is reported. The HP1451 construct consists of two domains similar to nucleic acid-binding domains. Two HP1451 molecules bind to the HP0525 NTDs on opposite sides of the hexamer, locking it in the closed form and forming a partial lid over the HP0525 chamber. From the structure, it is suggested that HP1451 acts as an inhibitory factor of HP0525 to regulate Cag-mediated secretion, a suggestion confirmed by results of in vitro ATPase assay and in vivo pull-down experiments.
Keywords: ATPase, crystal structure, Helicobacter pylori, type IV secretion, VirB11
Introduction
The spiral Gram-negative bacterial pathogen Helicobacter pylori is one of the world's most successful human pathogens, living in the stomach of up to half the world's population (Covacci et al, 1999). H. pylori has been shown to cause peptic and duodenal ulcers, mucosa-associated lymphoid tissue (MALT) lymphoma and gastric adenocarcinoma (Parsonnet et al, 1991). It has been classified by the world health authority as a category I carcinogen—a definite cancer-causing agent.
Different H. pylori strains display differing degrees of virulence due to their genetic diversity. The more virulent type I strains contain the Cag pathogenicity island, which is lacking in less virulent type II strains. The Cag pathogenicity island encodes a type IV secretion system (T4SS), which secretes the virulence factor CagA into host cells (Segal et al, 1999; Asahi et al, 2000; Backert et al, 2000; Odenbreit et al, 2000; Stein et al, 2000). After secretion, CagA is phosphorylated on tyrosine residues and causes extensions to appear on the host cell surface by interfering with the actin filament nucleation and polymerisation processes. Phosphorylated CagA also causes cell proliferation and scattering by interfering with cell signalling pathways, leading to activation of the mitogen-associated protein (MAP) kinase pathway (Segal et al, 1999; Censini et al, 2001; Mimuro et al, 2002). While many recent studies have focused on identifying the structure and mechanism of T4SSs, no regulatory mechanisms have so far been identified for the H. pylori Cag system.
T4SSs are one of five main secretion systems of Gram-negative bacteria. T4SSs are used by many pathogens to secrete effector molecules across their membranes into the cytoplasm of host cells or into the extracellular milieu. In addition to the stomach diseases associated with H. pylori, T4SSs are important in such diseases as whooping cough—caused by Bordetella pertussis (Weiss et al, 1993), crown gall tumours of plant stems—caused by Agrobacterium tumefaciens (Chilton et al, 1977), brucellosis—caused by Brucella species (O'Callaghan et al, 1999) and legionnaires disease—caused by Legionella pneumophila (Segal et al, 2005). T4SSs are also used for the transfer of plasmid DNA during conjugation, and are hence important in the transfer of genetic information and the spread of antibiotic resistance (Ochman et al, 2000).
The structure of T4SSs consists of a multiprotein assembly, which extends across the inner membrane, the periplasm and the outer membrane and, in some cases, includes a filamentous extracellular structure, the pilus. The 12 main constituent proteins of T4SSs are named VirB1 to VirB11 and VirD4, after their homologues in the archetypal T4SS of A. tumefaciens. Three of these proteins (VirB4, VirB11 and VirD4) are ATPases and are located in, or associated with, the inner membrane (Christie et al, 2005). It has been shown that, in A. tumefaciens, ATP hydrolysis by all three enzymes is required for transfer of the secretion substrate to the periplasmic T4SS components (Atmakuri et al, 2004).
As well as being essential for the A. tumefaciens T4SS, VirB11 homologues are also essential for protein transport by L. pneumophila, B. pertussis and H. pylori T4SSs, and most conjugation systems (Haase et al, 1995; Kotob and Burns, 1997; Rivas et al, 1997; Vogel and Isberg, 1999; Backert et al, 2000). A recent study has shown that inhibition of the Cag T4SS VirB11 homologue (named ‘HP0525') by selective inhibitor compounds resulted in a decrease in CagA transfer in vivo to undetectable levels (Hilleringmann et al, 2006). Previous structural studies of HP0525 have shown that this protein forms hexamers, each subunit consisting of two domains, an N-terminal domain (NTD) and a RecA-like C-terminal domain (CTD). Each domain of VirB11 forms a hexameric ring with the NTDs forming an open ring, sitting on top of the closed ring formed by the CTDs. Together these rings form a central chamber, sealed on the CTD side and open on the NTD side. The CTD ring has a grapple-like conformation, with two helices from each of the monomers pointing into the centre of the ring to form the claws of the grapple (Yeo et al, 2000). The nucleotide-binding site (NBS) is located between the two domains and nucleotide binding is essential for stabilising the NTD–CTD interface. Structures of HP0525 bound to nucleotides (or with sulphate mimicking the nucleotide phosphate) show NTDs in a closed state, while in the absence of nucleotide, the NTDs rotate outwards from the centre of the hexamer and this rotation is coupled with a disordering of the NTDs (Savvides et al, 2003). A later study of the VirB11 from Brucella suis (BsB11) showed that this homologue retained the hexameric double-ring structure of HP0525, but demonstrated a domain swap likely to be present in most other VirB11 homologues (Hare et al, 2006). These structural studies, as well as biochemical analysis of VirB11 homologues and T4SSs as a whole, have led to the suggestion that VirB11 acts as a chaperone in the unfolding of target proteins before secretion (Christie et al, 2005).
In a high-throughput yeast two-hybrid screen of the H. pylori proteome, it was shown that HP0525 interacts with a protein of unknown function, called HP1451 (Rain et al, 2001) (H. pylori proteins are herein referred to as HPXXXX, where XXXX indicates open reading frame number, as in the H. pylori genome sequence; Tomb et al, 1997). Furthermore, the interacting region of HP1451 was approximately mapped to residues 92–264. The interaction was confirmed by a second study, wherein a stable complex of HP0525 and a construct of HP1451 consisting of residues 92–264 was purified (Terradot et al, 2004). HP1451 has no known function and is located on the chromosome distant from the Cag pathogenicity island. It shows weak homology to the SpoIIIJ-Associated Gene (JAG) of Bacillus subtilis, a gene co-transcribed with SpoIIIJ. While JAG itself has no proposed role, SpoIIIJ is a sporulation factor that has been suggested to act as a protein translocase due to its transmembrane helices and homology with the Escherichia coli protein translocase YidC (Murakami et al, 2002).
In the study presented here, we have determined the crystal structure of the HP0525-interacting domain of HP1451 bound to HP0525. This complex suggests a function for HP1451 as a regulator of HP0525 ATPase activity, a role confirmed by ATPase activity measurements and in vivo pull-down experiments. This is the first regulator of VirB11 activity ever identified, and its structure bound to its target ATPase suggests a novel mode of action.
Results and discussion
Identification of a minimal domain of HP1451 interacting with HP0525
After crystallisation attempts using the originally identified HP0525-binding region of HP1451 (HP1451C1, residues 92–274) failed to produce diffraction quality crystals, further HP1451 constructs were prepared to narrow down or expand the interacting region and to attempt crystallisation. These constructs were (see Supplementary Figure 1) the following: full-length HP1451 (HP1451F), residues 113–274 (HP1451C2) and residues 145–264 (HP1451C3). HP1451C2 is slightly shorter than HP1451C1, with a sequence of low complexity removed, and begins at the N-terminus of a predicted α-helix. HP1451C3 includes the C-terminal region of HP1451 that corresponds to the most homologous region of the JAG protein from B. subtillis (Supplementary Figure 1). The NTD (residues 1–136) and CTD (residues 137–330) of HP0525 were also cloned separately to attempt to characterise the HP1451-binding region on HP0525.
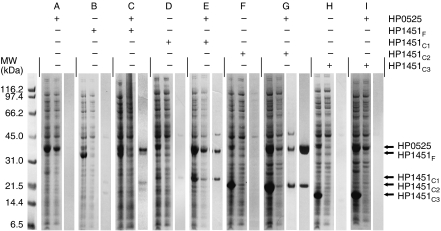
All HP1451 constructs are insoluble when expressed alone in E. coli cells, with the exception of HP1451C1, which is slightly soluble (Figure 1, sections B, D, F and H). Of the two fragments of HP0525, only one (the CTD) stably expressed on its own (results not shown). HP1451F, HP1451C1 and HP1451C2 are soluble when coexpressed with full-length HP0525, and form stable complexes with HP0525 (Figure 1, sections C, E and G). Thus, HP1451F, HP1451C1 and HP1451C2 exhibit increased solubility as a result of complex formation with HP0525. However, these proteins remain poorly soluble when coexpressed with just the CTD of HP0525 (results not shown), indicating that the CTD of HP0525 is not the region of HP0525 that interacts with HP1451. HP1451C3 is not soluble when coexpressed with any HP0525 construct and therefore presumably does not form a complex with HP0525 (Figure 1, section I). From these results it appears that the HP0525/HP1451 interaction requires the NTD of HP0525 and residues 113–144 of HP1451.
Figure 1.
Expression, solubility and purification of HP1451 constructs and HP0525/HP1451 complexes. Sections A–I correspond to the expression and Talon purification of different His-tagged constructs of HP1451 both alone and when coexpressed with HP0525. The clones used in expression are indicated with + and − in the table above the gel. For each section, the first lane contains the induced whole-cell extract, the second lane shows the soluble proteins after lysis and clarification, and the third lane shows the elution fraction from the Talon column. Section G contains a fourth lane that corresponds to the final purified HP0525/HP1451C2 complex after subsequent purification steps. HP1451 constructs are not soluble when expressed alone; however, HP1451F, HP1451C1 and HP1451C2 show a dramatic increase in solubility when coexpressed with HP0525.
HP0525/HP1451F and HP0525/HP1451C2 complexes were purified and crystal trials were attempted for both. The HP0525/HP1451C2 complex could be produced stably, in abundance, and led to the production of crystals diffracting to 2.4 Å. This was not the case for HP0525/HP1451F where we observed a rapid breakdown of HP1451.
Determination of the structure of the HP0525/HP1451C2 complex
Originally, clusters of plate-shaped crystals were grown against a solution containing 0.1 M MES pH 6.0, 8% (w/v) PEG 8000; these diffracted to 3.2 Å. Adding 10 mM EDTA to the crystallisation solution yielded rectangular three-dimensional crystals that diffracted to 2.4 Å. These improved crystals are in P1 spacegroup, with unit-cell dimensions of a=86.85 Å, b=86.90 Å, c=104.13 Å, α=111.04°, β=95.98° and γ=104.94°.
From SDS–PAGE bands of the purified complex it appears that the ratio of HP0525:HP1451C2 in the complex is 6:1 or 6:2 (Figure 1, section G). The unit-cell dimensions suggest the presence of an HP0525 hexamer in the unit cell (Matthew's coefficient 3.07 for six HP0525 molecules), with some space available for one or two molecules of HP1451C2. Molecular replacement (MR) using the nucleotide-free (open) HP0525 hexameric structure (PDB code: 1NLZ) as a search model failed. However, MR using the ADP-bound (closed) form of the HP0525 hexamer (PDB code: 1G6O) as a search model yielded a clear solution. An electron density map calculated using phases derived from this MR solution showed clear density for two HP1451C2 molecules. The final model (R and Rfree factors of 22.9 and 26.9%, respectively, with good stereochemistry) contains residues 115–259 of two chains of HP1451C2 (chains G and H) and a hexamer of HP0525 (chains A–F; Supplementary Table 1). The NBS of all six HP0525 chains are unoccupied.
Structure of HP1451C2
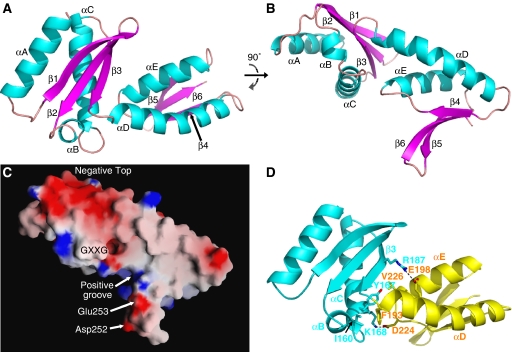
HP1451C2 consists of two domains, the first from residue 115 to 191 and the second from 192 to 259 (Figure 2). The first domain is made of three β-strands (β1–3) forming a mixed β-sheet and three α-helices (αA–C), two of which pack against the β-sheet, with the third at one of the sheet's ends. The secondary structure elements are arranged in an α–β–β–α–α–β topology and the fold is the KH-domain fold, commonly found in nucleic acid-binding proteins. The signature sequence of KH-domains is VIGXXGXXI (Grishin, 2001). In HP1451, this characteristic sequence is slightly different (159-LIGEKGYRY-167), but produces the same structural element (i.e., the turn between αB and αC; see also surface in Figure 2C) and contains the two absolutely conserved glycines capping the two helices. This turn has been shown to interact with RNA in the KH-domains of NusA (an essential bacterial transcription factor) and NOVA (a neuronal splicing factor) (Lewis et al, 2000; Beuth et al, 2005).
Figure 2.
Cartoon and surface representations of the HP1451C2 monomer. (A, B) Two views of HP1451C2 rotated 90° about the x-axis, termed top view (A) and side view (B). These orientations are used in subsequent figures. α-Helices are coloured cyan, β-strands coloured magenta and loops coloured pink. Secondary structure elements are labelled as in Supplementary Figure 1. (C) Surface representation of the HP1451C2 monomer with electrostatic potential indicated by red for the most negative regions and blue for the most positive regions. Regions and residues highlighted in the text are indicated. View is as in panel B. (D) Interactions between the two domains of HP1451C2. View is as in panel A and colour-coding is KH-domain in cyan and S8 domain in yellow. Side chains of the residues of the interface are shown as stick representation and labelled. Colours for the stick representation are cyan for KH-domain carbon, yellow for S8-domain carbon, red for oxygen and blue for nitrogen. All the panels, except panel C (panel C was produced using GRASP; Nicholls et al, 1991), were produced using Pymol (DeLano, 2002).
The second domain looks similar to the KH-domain in that it has a three-stranded β-sheet (β4–6) packed against two α-helices (αD–E). However, this domain differs in that (i) the β-sheet is antiparallel, (ii) there is no short helix and (iii) the topology is α–β–α–β–β. In both the domains, the β-sheet is twisted so that two of the strands point away from the helices at one end, a feature that is more marked in the second domain (Figure 2A and B). The short helix and the GXXG motif that are characteristic of KH-domains are missing in this second domain. This domain is structurally homologous to other nucleic acid-binding proteins such as the NTD of ribosomal protein S8 and DNaseI.
The top surface of HP1451C2 is largely negatively charged (‘negative top' in Figure 2C), while the surface facing the central chamber of HP0525 is mainly positive (Figure 2C). The exception to this is the patch of negative charge caused by Asp252 and Glu253, which is important for the interaction with HP0525 (Figure 2C, see below). The positive area on the underside of HP1451 is contributed by the side chains of Lys168, Lys221 and Lys254 and forms a groove between the strands β5 and β6 and the main body of the structure (‘positive groove' in Figure 2C).
The two HP1451 domains (hereafter referred to as the KH-domain and S8-domain) interact with each other via two salt bridges involving residues Lys168:Asp224 and Arg187:Glu198. Interactions are also established through the insertion of Val226 at the N-terminal end of helix αE into a hydrophobic pocket of the KH-domain, and through hydrophobic interactions between Phe193, next to the link between the two domains, and Tyr167 and Ile160, of the αB and αC helices, respectively (Figure 2D).
Structure of the HP0525/HP1451C2 complex
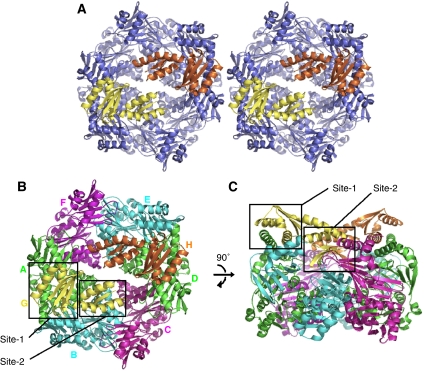
Two HP1451C2 molecules are positioned on the NTD side of the HP0525 hexameric ring opposite each other, so that each HP1451C2 molecule has its KH-domain at the outside and its S8-domain pointing into the centre of the ring (Figure 3). The HP1451C2 molecules cap the HP0525 cavity, creating a chamber completely sealed at its base by the HP0525 CTDs, but with gaps in the top between the HP1451C2 and HP0525 molecules.
Figure 3.
Cartoon representation of HP0525/HP1451C2 complex. (A) Stereoview of the complex, with the HP0525 hexamer coloured blue and HP1451C2 chains coloured yellow and orange. (B, C) Top and side views, respectively, of the HP0525/HP1451C2 complex. HP0525 chains A and D are coloured green, chains B and E are cyan and chains C and F are magenta. HP1451C2 monomers are coloured yellow for chain G and orange for chain H. The two main sites of interaction mentioned in the text are indicated by the black box outlines and are labelled. Orientations in panels B and C are as in Figure 2A and B, respectively. All the panels were produced using Pymol (DeLano, 2002).
Each HP1451C2 molecule interacts primarily with the NTDs of two adjacent monomers of HP0525 and also interacts weakly with a third HP0525 monomer and some residues of the CTDs (Figures 3 and 4). Here we describe the structure referring to the HP0525 chains as A to F (when the ring is viewed from the NTD side the monomers are numbered anticlockwise) and the HP1451C2 chains as G and H (Figure 3B). Chain G interacts mainly with HP0525 chains A and B and interacts weakly with chain C, while HP1451C2 chain H has its strongest interactions with chains D and E and a weak interaction with chain F. The following discussion concentrates on chain G of HP1451C2 and chains A, B and C of HP0525; however, the interactions are reproduced in the other half of the structure, as NCS restraints were used during refinement (the NCS root mean square deviation (r.m.s.d.) for chains G and H is 0.15 Å; for chains A and D is 0.04 Å; for B and E is 0.04 Å and for chains C and F is 0.05 Å).
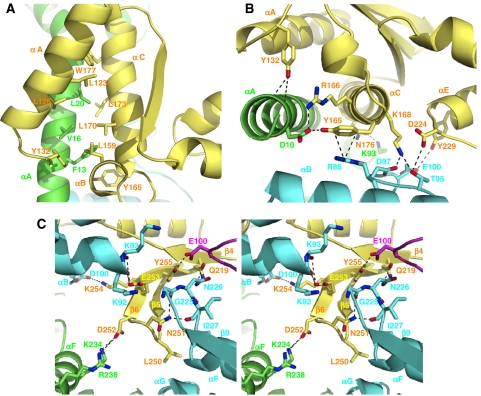
Figure 4.
Close-up images of the HP0525/HP1451C2 interface. In all the panels the cartoon backbone is coloured as in Figure 3B: yellow, HP1451 chain G; green, HP0525 chain A; cyan, HP0525 chain B and magenta, HP0525 chain C. Side chains important in the interface are shown as sticks, with the carbon atoms coloured according to chain (as cartoon), nitrogen coloured blue and oxygen coloured red. The side chains are labelled in the colour corresponding to their chain. (A) Top view (as Figure 2A) of the interaction at site-1, where helix αA of HP0525 chain A interacts with helices αA, αB and αC of HP1451C2. Side chains of residues making hydrophobic contacts along the length of the helices are shown. (B) Side view (as Figure 2B) of the interaction at site-1 with residues making hydrophilic interactions indicated. (C) Side stereoview (as in Figure 2B) of the interactions at site-2, where HP1451C2 residues make contact with residues from three different chains of HP0525. All the panels were produced using Pymol (DeLano, 2002).
HP1451C2 has two main sites where it interacts with HP0525; the first (site-1) involves principally helices αA and αC of the KH-domain and the second (site-2) involves the β5–β6 loop of the S8-domain (Figure 3).
At site-1, helices αA and αC of HP1451C2 interact with helix αA of HP0525 chain A and make hydrophobic, side chain–side chain interactions along most of the length of the helices. Leu159 of HP1451C2 helix αB also forms a part of this internal hydrophobic region (Figure 4A). On one end of this helical hydrophobic region, the interactions become polar with HP1451C2 residues Tyr132 and Tyr165 making hydrophilic contacts with HP0525 helix αA, Tyr132 with backbone carbonyls and Tyr165 with the side chain of HP0525 Asp10 (Figure 4B). There is a charge–charge interaction between HP1451C2 Arg166 and HP0525 Asp10, and finally the π-electron cloud of HP1451 Tyr165 interacts with Arg86 of HP0525 chain B. The underside of helix αC of HP1451C2 also interacts with HP0525; here HP1451C2 Asn176 interacts with Lys93 of the αB–β4 loop of HP0525 chain A. Two residues from the S8-domain also play a role at the interface at site-1: Asp224 of the HP1451C2 β4–αE loop and Tyr229 of helix αE interact with HP0525 chain B Thr95 and Glu100, respectively (Figure 4B).
At site-2, the β5 and β6 strands of the S8-domain insert down into the HP0525 structure at the interface between the NTD and CTD of molecule B. This β-hairpin is pointing towards, but falling approximately 20 Å short of, the NBS. This part of HP1451C2 makes interactions with residues in several different locations of HP0525, namely helix αF of molecule A, the β9–αF loop, the N-terminal end of helix αG and the αB–β4 loop of molecule B, and the αB–β4 loop of molecule C (Figure 4C). This is the only site of interaction between HP1451C2 and molecule C of HP0525 and involves HP1451C2 Tyr255 and HP0525 Glu100. Site-2 consists almost entirely of polar contacts with only HP1451C2 Leu250 forming hydrophobic interactions. There is a very significant charge–charge interface involving the acidic residues Asp252 and Glu253 of HP1451C2 and the basic residues Lys92, Lys93, Lys234 and Arg238 of HP0525 (Figures 2C and 4C). The carboxylic acid group of Asp252 forms a salt bridge with Lys234 and is also close to the guanidinium group of Arg238 both on HP0525 molecule B. Glu253 is close to Lys92 and Lys93 of HP0525 chain B, but interacts with the backbone atoms rather than with the charge groups of the side chains. Another important HP1451C2 residue at this interface is Asn251, which forms close polar interactions with backbone oxygen atoms of residues 225 and 227 of the β9–αF loop of HP0525 molecule B (Figure 4C).
It was observed that the integrity of the complex is compromised in high-salt buffers to such an extent that 0.5 M NaCl led to a complete dissociation of HP1451C2 from HP0525 (see below). This suggests that the polar interactions, rather than the hydrophobic helix–helix interactions at site-1, are the predominant interactions in maintaining the complex.
Effect of HP1451 binding on the structure of HP0525
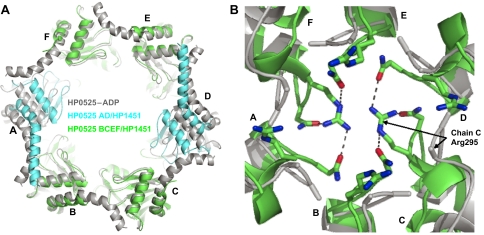
HP1451C2 binding to HP0525 induces two notable changes in the HP0525 structure. First, HP1451C2 locks the HP0525 NTDs into the closed state, whereas in the structure of unliganded HP0525 alone, the NTDs are flexible. The closed state was previously observed only in nucleotide-bound forms of the structure or when a sulphate ion from the crystallisation conditions binds in the β-phosphate position of the NBS. In the HP1451-bound structure, the HP1451 pins the NTDs of chains A and B (or D and E) together and rigidly links them to the CTD. Since the interaction of HP1451 with chains C and F is small, these subunits might be less securely locked. This is reflected in the structure by the lower average B-values of chains A, B, D, and E NTDs compared to those of chain C and F (chains A/D—30.5/30.6, chains B/E—32.4/32.4 and chains C/F—43.7/43.8 (residues 22–136 used for calculations)). In the HP1451-bound state, nucleotide access to the NBS of chains A, B, D and E would be dramatically reduced. In fact the NTDs of HP0525 monomers A and D are positioned closer to the central chamber than in the nucleotide-bound forms of the protein. These NTDs are pulled into this ‘more closed' state because of the interaction of the αA helices with HP1451C2; this interaction also leads to a straightening of the αA helices (Figure 5A).
Figure 5.
A comparison of the HP0525–ADP structure with that of HP0525 in complex with HP1451C2. In both the panels the HP0525–ADP structure is coloured grey and the monomers of HP0525 are labelled A–F. (A) Overall view of the conformational changes occurring upon HP1451 binding. The NTDs of HP0525 are displayed from the same view as in Figure 3A and B. The greatest domain movement is of the NTDs of chains A and D, and these are coloured cyan for the HP0525/HP1451C2 structure, while the other chains are green. (B) Grapple of HP0525 from the same angle as A. Here all monomers of the HP0525/HP1451C2 complex are coloured green. The side chains of residues Arg295 and Asn296 are shown as stick representations for both structures but only the Cβ atoms are visible for the HP0525–ADP complex as the rest of the side chains are disordered. All the panels were produced using Pymol (DeLano, 2002).
In all previous structures of HP0525 (and indeed in the homologue from B. suis) there is a small (10 Å) hole in the centre of the CTD grapple (Yeo et al, 2000; Savvides et al, 2003; Hare et al, 2006). In the complex with HP1451C2, the side chains of Arg295 and Asn296 at the tip of the grapple are rearranged to form a plug in this hole. In previous structures, these side chains were disordered, but in the HP0525/HP1451C2 complex, Arg295 of chains C and F point across the hole blocking it and Asn296 from all chains stabilises the positive charge of the plug (Figure 5B). Other potential stabilizing factors could originate from long-range structural effects induced by HP1451-binding or indirect effects of crystal packing (however, note that no direct crystal packing is observed at or around the plug).
ATPase activity
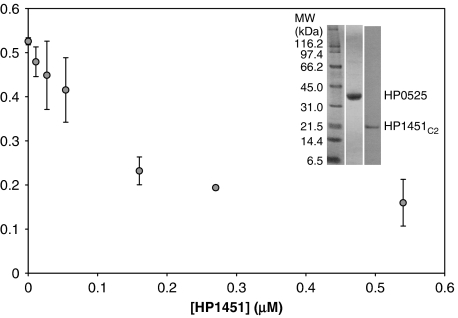
Flexibility of the HP0525 NTDs is probably important for nucleotide binding and release as the NBS lies between the two domains. The loss of NTD flexibility in the HP0525/HP1451C2 structure suggests that HP1451 might affect ATPase activity. Thus, an ATPase assay was set up and the effect of increasing concentration of HP1451C2 on HP0525 ATPase activity was monitored. Pure untagged HP0525 was obtained as described in Materials and methods. HP1451C2 is insoluble when expressed alone in E. coli; therefore, purification of HP1451C2 for the ATPase assay required expression of the HP0525/HP1451C2 complex, binding of the complex via the His6 tag of HP1451C2 to Talon resin, and dissociating the complex and eluting HP0525 using 0.5 M NaCl. Under such conditions, HP1451C2 alone remains bound to the column. HP1451C2 was then eluted using imidazole and further purified using gel filtration, also at 0.5 M NaCl (Figure 6, inset).
Figure 6.
ATPase activity of HP0525 in the presence of various concentrations of HP1451C2. Reactions were carried out as described in Materials and methods. Error bars show the standard deviation of the data. The inset demonstrates the purity of the proteins used in the experiment. Increasing HP1451C2 concentration leads to a decrease in HP0525 activity to approximately 30% of its original rate once a 1:1 molar ratio of HP1451C2 is added.
ATPase activity of HP0525 was measured using the ATPase assay (high sensitivity) kit from Innova Biosciences. Thirty-minute reactions were carried out and the final phosphate concentrations was measured (see Materials and methods). HP1451C2 was added at various concentrations (0–0.54 μM) to 0.25 μM HP0525. The maximum inhibition exerted by HP1451C2 is reached at a molar ratio of approximately 1:1 of HP0525:HP1451C2 (i.e., a threefold molar excess of HP1451C2 for formation of 6:2 complexes). This inhibition caused ATP hydrolysis by HP0525 to drop to 30% of its native value (Figure 6), indicating that four of the six ATPase subunits are inhibited by HP1451. This result is consistent with the observation that HP1451C2 interacts primarily with four of the six HP0525 subunits, locking them in the closed conformation. The same assay repeated with B. suis VirB11 showed no inhibitory effect of HP1451C2, demonstrating the specificity of HP1451 for HP0525 (results not shown).
In vivo pull-down experiments
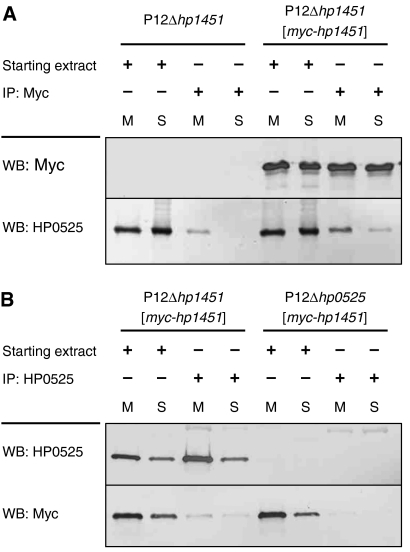
To demonstrate an interaction between HP0525 and HP1451 in H. pylori, we complemented an isogenic Δhp1451 mutant of strain P12 with plasmid pWS278, from which a myc-tagged version of hp1451 is expressed. Neither deletion of hp1451 nor complementation with myc-hp1451 caused any interference with HP0525 production or the function of the Cag T4SS (data not shown). Since HP0525 is found both as a soluble protein and in association with the cytoplasmic membrane (Pattis et al, 2007), we fractionated H. pylori cells into soluble and membrane fractions and performed anti-Myc and anti-HP0525 immunoprecipitations from both fractions separately. Although some HP0525 coprecipitated unspecifically with anti-Myc antibodies in the absence of Myc-HP1451 from the membrane fraction, coprecipitation of HP0525 with Myc-HP1451 from both membrane and soluble fractions demonstrated an interaction (Figure 7A; compare fourth and eighth lanes). In the reverse experiment, co-immunoprecipitation of Myc-HP1451 with HP0525 from both fractions demonstrated a specific, albeit weak, interaction between the two proteins (Figure 7B). Thus, HP1451 and HP0525 interact in vivo.
Figure 7.
In vivo pull-down experiments. (A) A Myc-tagged version of HP1451 was pulled down by anti-Myc immunoprecipitation (IP) from membrane (M) and soluble (S) fractions of H. pylori strain P12Δhp1451[myc-hp1451]. As a control, the same immunoprecipitation was performed from the mutant P12Δhp1451, which does not express myc-hp1451. Co-immunoprecipitation of HP0525 was determined by western blot (WB) using anti-HP0525. (B) In a reciprocal experiment, HP0525 was immunoprecipitated from membrane and soluble fractions of strains P12Δhp1451[myc-hp1451] and P12Δhp0525[myc-hp1451], and co-immunoprecipitation of Myc-HP1451 was determined by western blot.
Conclusion
HP1451 was initially discovered in a protein–protein interaction screen of the H. pylori proteome, and the interaction with HP0525 was subsequently confirmed biochemically (Rain et al, 2001; Terradot et al, 2004). However, no function for HP1451 was ascribed and sequence homology analysis suggested a role in RNA metabolism, not T4SS. Here we provide the additional insights that the two proteins interact in vivo and that HP1451 modulates the ATPase activity of HP0525. We also describe the mechanism by which HP1451 accomplishes its function. Although a fragment of HP1451 was used for the structural and biochemical work, this fragment is likely to recapitulate the function of full-length HP1451 as complexes of full-length HP1451 and HP0525 could be produced and the full-length HP1451 was pulled down with HP0525 in the in vivo pull-down experiments reported here (Figures 1 and 7).
Several functions of HP1451 can be suggested from the HP1451C2/HP0525 complex structure. First, the observation that each HP1451 molecule interacts with three HP0525 molecules, and that the two HP1451 molecules appear to lock the HP0525 hexamer in its closed state, suggests a role for HP1451 in modulating HP0525 ATPase activity. This is confirmed by the ATPase assay results presented here. In the A. tumefaciens T4SS, it has been shown that ATP hydrolysis by VirB11 is essential for the transfer of substrate to the inner membrane channel components. This strongly suggests that HP1451 is acting as an inhibitor of Cag-mediated secretion by blocking HP0525 ATPase activity. Second, the location of the HP1451 interaction site on HP0525 could be significant. HP1451 indeed obstructs the chamber formed by the two domains of HP0525. As it has been hypothesised that the substrate of the Cag T4SS, CagA, might pass through the chamber (Yeo et al, 2000), a possible role of HP1451 is to block transport. Third, it has been shown that in A. tumefaciens it is the NTDs of VirB11 that are responsible for interaction with the other T4SS components (Ward et al, 2002). Therefore, HP1451 could prevent the protein–protein interactions that appear to be important for the function of VirB11 homologues. Finally, another possibility is that HP1451 affects the membrane localisation of HP0525; however, this is unlikely as HP1451 binds to the internal hydrophilic cavity formed by the HP0525 hexamer, not the more hydrophobic external surface which presumably interact with the membrane.
In summary, the HP0525/HP1451 structure provides an intriguing glimpse at the first identified negative regulatory mechanism of a T4SS. Knocking out HP1451 does not prevent secretion through the Cag system, consistent with a negative rather than positive regulatory function as predicted from the structure and ATPase assay. Unfortunately, due to difficulty in measuring the native expression level of HP1451 in H. pylori, it has not been possible to assess the effect of over-expression. However, a study showing that small-molecule inhibitors of HP0525 ATPase activity prevent CagA translocation in vivo provides a firm foundation for believing that protein-mediated inhibition of HP0525 would have the same affect (Hilleringmann et al, 2006).
Materials and methods
Plasmids and constructs
For more information on the above, see Supplementary data.
Interaction studies
HP0525 and HP1451 constructs were transformed individually, and cotransformed in all possible combinations into BL21 star (DE3) cells (Invitrogen), using ampicillin and spectinomycin to select for pET151 and pCDF vectors, respectively. Cultures (0.5 l) of transformed cells were grown with shaking at 37°C to an A600 of 0.4 before being moved to 20°C. After 1 h at 20°C, isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and the cultures were incubated for a further 16 h. The cells were harvested by centrifugation, resuspended in 40 ml Buffer A (20 mM Tris/HCl pH 7.6, 150 mM NaCl, 10% (v/v) glycerol) and frozen at −20°C.
All subsequent steps were carried out at 4°C. The cells were defrosted and one tablet of Protease inhibitor cocktail EDTA free (Roche) was added. After cell lysis by homogenisation and DNA fragmentation by sonication, the lysate was clarified by centrifugation at 18 000 r.p.m. for 30 min in a Sorvall SS-34 rotor. The clarified lysate was loaded onto a chromatography column containing 20 ml Talon resin (Clontech), and equilibrated in buffer A. The resin was washed with 100 ml buffer A and then with 50 ml of 96% buffer A plus 4% buffer B (20 mM Tris/HCl pH 7.6, 150 mM NaCl, 10% (v/v) glycerol, 0.5 M imidazole). Finally the proteins still bound to the column were eluted in a gradient from 4% to 100% buffer B over 100 ml.
Large-scale expression and purification
For more information on the above procedures, see Supplementary data.
Crystallisation, data collection and processing
Pure HP0525/HP1451C2 was concentrated to 7 mg/ml and crystallised at 4°C by hanging drop, vapour diffusion in drops containing 1 μl protein and 1 μl precipitant suspended over a reservoir containing 100 μl of 100 mM MES pH 6.0, 8% polyethyleneglycol (PEG) 8000, 10 mM ethylenediaminetetraacetic acid (EDTA). After 2 days rectangular crystals appeared, which after a week's growth reached a size of 200 × 100 × 100 μm. The crystals were cryoprotected by transferring to drops with gradually increasing glycerol concentration to a final concentration of 25% (v/v), and flash frozen in liquid nitrogen. Data were collected to 2.4 Å under a cryostream of nitrogen gas at 100K at ESRF beamline ID29 (Supplementary Table 1). The data were integrated using MOSFLM (Leslie, 1992) and merged using SCALA (Evans, 1993) part of the CCP4 programme suite (CCP4, 1994). Phases were obtained by MR, using the programme MOLREP and a hexamer of HP0525–ADP as a search model (PDB code 1G6O) (Vagin, 1997). All refinement was carried out using the programme REFMAC (Murshudov et al, 1997). The programme Coot was used for model building (Emsley and Cowtan, 2004). Initial rigid-body refinement of the 12 HP0525 domains allowed density for two HP1451C2 molecules to be visualised. The HP1451C2 chains were manually built, initially as poly-alanine, with side chains being added as density became visible. Multiple rounds of building iterated with restrained refinement led to a final model with an Rfactor of 22.99% and an Rfree of 26.97% and good geometry (Supplementary Table 1).
ATPase assay
The Innova Biosciences ATPase assay (high sensitivity) kit was used following the manufacturer's instructions. ATPase reactions were carried out for 30 min at room temperature in 100 mM Tris pH 7.4, 1 mM ATP and 5 mM Mg2+. HP0525 (0.25 μM) was used with 0–0.54 μM HP1451C2. Protein concentrations were determined using the extinction coefficient at 280 nm as calculated by amino-acid analysis carried out by the PNAC facility at the University of Cambridge.
In vivo pull-down experiments
Bacteria grown on agar plates were suspended in PBS and washed twice. An amount of 5 × 1010 bacterial cells was resuspended in preparation buffer (50 mM Tris–HCl, pH 8.0, 1 mM PMSF, 1 μM leupeptin, 1 μM pepstatin) and the cells were lysed by sonication. Unbroken cells were removed by centrifugation for 10 min at 10 000 g. The supernatant was separated by ultracentrifugation (45 min, 230 000 g) into soluble (cytoplasmic and periplasmic) and total membrane fractions. Membranes were carefully resuspended in radioimmunoprecipitation (RIPA) buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 0.25% sodium deoxycholate, 1 mM PMSF, 10 μg/ml leupeptin, 10 μg/ml pepstatin) and the soluble fraction was adjusted to the same composition by addition of NaCl, EDTA, detergents and protease inhibitors. To remove unspecifically interacting proteins, the lysates were incubated with prewashed protein G-agarose (Roche Diagnostics) for 2 h at 4°C and centrifuged. To the supernatants, 5 μl of the appropriate antiserum was added and samples were incubated for 3 h at 4°C. Then, 50 μl of prewashed protein G-agarose was added and samples were incubated at 4°C for an additional 2 h. After three washing steps with RIPA buffer, proteins were eluted with 100 mM glycine, pH 2.7, or by boiling in SDS–PAGE sample solution.
Coordinates
The atomic coordinates and diffraction data in the form of structure factor amplitudes have been deposited in the Protein Data Bank under ID code 2PT7.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by grant 065932 from the Wellcome Trust (to GW), by MRC studentships to SH and RW, a Royal Society University Research Fellowship to RB and by grants Fi 953/1-1 and Ha 2697/9-1 from the Deutsche Forschungsgemeinschaft (to WF and RH, respectively).
References
- Asahi M, Azuma T, Ito S, Ito Y, Suto H, Nagai Y, Tsubokawa M, Tohyama Y, Maeda S, Omata M, Suzuki T, Sasakawa C (2000) Helicobacter pylori CagA protein can be tyrosine phosphorylated in gastric epithelial cells. J Exp Med 191: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmakuri K, Cascales E, Christie PJ (2004) Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol 54: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backert S, Ziska E, Brinkmann V, Zimny-Arndt U, Fauconnier A, Jungblut PR, Naumann M, Meyer TM (2000) Translocation of the Helicobacter pylori CagA protein in gastric epithelial cells by a type IV secretion apparatus. Cell Microbiol 2: 155–164 [DOI] [PubMed] [Google Scholar]
- Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA (2005) Structure of a Mycobacterium tuberculosis NusA–RNA complex. EMBO J 24: 3576–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP 4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Censini S, Stein M, Covacci A (2001) Cellular responses induced after contact with Helicobacter pylori. Curr Opin Microbiol 4: 41–46 [DOI] [PubMed] [Google Scholar]
- Chilton MD, Drummond MH, Merio DJ, Sciaky D, Montoya AL, Gordon MP, Nester EW (1977) Stable incorporation of plasmid DNA into higher plant cells: the molecular basis of crown gall tumorigenesis. Cell 11: 263–271 [DOI] [PubMed] [Google Scholar]
- Christie PJ, Atmakuri K, Krishnamoorthy V, Jakubowski S, Cascales E (2005) Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu Rev Microbiol 59: 451–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covacci A, Telford JL, Del Giudice G, Parsonnet J, Rappuoli R (1999) Helicobacter pylori virulence and genetic geography. Science 284: 1328–1333 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System. San Carlos, CA, USA: DeLano Scientific, http://www.pymol.org [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Evans PR (1993) Data reduction. Proceedings of CCP4 Study Weekend, 1993, on Data Collection & Processing 114–122. Warrington, UK: Daresbury Laboratory [Google Scholar]
- Grishin NV (2001) KH domain: one motif, two folds. Nucleic Acids Res 29: 638–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase J, Lurz R, Grahn AM, Bamford DH, Lanka E (1995) Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol 177: 4779–4791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Bayliss R, Baron C, Waksman G (2006) A large domain swap in the VirB11 ATPase of Brucella suis leaves the hexameric assembly intact. J Mol Biol 360: 56–66 [DOI] [PubMed] [Google Scholar]
- Hilleringmann M, Pansegrau W, Doyle M, Kaufman S, MacKichan ML, Gianfaldoni C, Ruggiero P, Covacci A (2006) Inhibitors of Helicobacter pylori ATPase Cagalpha block CagA transport and cag virulence. Microbiology 152: 2919–2930 [DOI] [PubMed] [Google Scholar]
- Kotob SI, Burns DL (1997) Essential role of the consensus nucleotide-binding site of PtlH in secretion of pertussis toxin from Bordetella pertussis. J Bacteriol 179: 7577–7580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW (1992) Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4+ESF-EAMCB Newsletter on Protein Crystallography, Vol. 26. Warrington, UK: Daresbury Laboratory [Google Scholar]
- Lewis HA, Musunuru K, Jensen KB, Edo C, Chen H, Darnell RB, Burley SK (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile × syndrome. Cell 100: 323–332 [DOI] [PubMed] [Google Scholar]
- Mimuro H, Suzuki T, Tanaka J, Asahi M, Haas R, Sasakawa C (2002) Grb2 is a key mediator of helicobacter pylori CagA protein activities. Mol Cell 10: 745–755 [DOI] [PubMed] [Google Scholar]
- Murakami T, Haga K, Takeuchi M, Sato T (2002) Analysis of the Bacillus subtilis spoIIIJ gene and its Paralogue gene, yqjG. J Bacteriol 184: 1998–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D 53: 240–255 [DOI] [PubMed] [Google Scholar]
- Nicholls A, Sharp KA, Honig B (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11: 281–296 [DOI] [PubMed] [Google Scholar]
- O'Callaghan D, Cazevieille C, Allardet-Servent A, Boschiroli ML, Bourg G, Foulongne V, Frutos P, Kulakov Y, Ramuz M (1999) A homologue of the Agrobacterium tumefaciens VirB and Bordetella pertussis Ptl type IV secretion systems is essential for intracellular survival of Brucella suis. Mol Microbiol 33: 1210–1220 [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–304 [DOI] [PubMed] [Google Scholar]
- Odenbreit S, Puls J, Sedlmaier B, Gerland E, Fischer W, Haas R (2000) Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Parsonnet J, Friedman GD, Vandersteen DP, Chang Y, Vogelman JH, Orentreich N, Sibley RK (1991) Helicobacter pylori infection and the risk of gastric carcinoma. N Engl J Med 325: 1127–1131 [DOI] [PubMed] [Google Scholar]
- Pattis I, Laugks E, Haas R, Fischer W (2007) The Helicobacter pylori CagF protein is a type IV secretion chaperone-like molecule that binds close to the C-terminal secretion signal of the CagA effector protein. Microbiology 153: 2896–2909 [DOI] [PubMed] [Google Scholar]
- Rain JC, Selig L, De Reuse H, Battaglia V, Reverdy C, Simon S, Lenzen G, Petel F, Wojcik J, Schachter V, Chemama Y, Labigne A, Legrain P (2001) The protein–protein interaction map of Helicobacter pylori. Nature 409: 211–215 [DOI] [PubMed] [Google Scholar]
- Rivas S, Bolland S, Cabezon E, Goni FM, de la Cruz F (1997) TrwD, a protein encoded by the IncW plasmid R388, displays an ATP hydrolase activity essential for bacterial conjugation. J Biol Chem 272: 25583–25590 [DOI] [PubMed] [Google Scholar]
- Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, Lanka E, Buhrdorf R, Fischer W, Haas R, Waksman G (2003) VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J 22: 1969–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal ED, Cha J, Lo J, Falkow S, Tompkins LS (1999) Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc Natl Acad Sci USA 96: 14559–14564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal G, Feldman M, Zusman T (2005) The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev 29: 65–81 [DOI] [PubMed] [Google Scholar]
- Stein M, Rappuoli R, Covacci A (2000) Tyrosine phosphorylation of the Helicobacter pylori CagA antigen after cag-driven host cell translocation. Proc Natl Acad Sci USA 97: 1263–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradot L, Durnell N, Li M, Li M, Ory J, Labigne A, Legrain P, Colland F, Waksman G (2004) Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol Cell Proteomics 3: 809–819 [DOI] [PubMed] [Google Scholar]
- Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A et al. (1997) The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388: 539–547 [DOI] [PubMed] [Google Scholar]
- Vagin A (1997) MOLREP: an automated program for molecular replacement. J Appl Cryst 30: 1022–1025 [Google Scholar]
- Vogel JP, Isberg RR (1999) Cell biology of Legionella pneumophila. Curr Opin Microbiol 2: 30–34 [DOI] [PubMed] [Google Scholar]
- Ward DV, Draper O, Zupan JR, Zambryski PC (2002) Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci USA 99: 11493–11500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss AA, Johnson FD, Burns DL (1993) Molecular characterization of an operon required for pertussis toxin secretion. Proc Natl Acad Sci USA 90: 2970–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo HJ, Savvides SN, Herr AB, Lanka E, Waksman G (2000) Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV secretion system. Mol Cell 6: 1461–1472 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data