Abstract
Infection with human papillomaviruses (HPV) is strongly associated with the development of cervical cancer. The HPV E6 oncogene induces apoptosis in cervical cancer precursor lesions but the mechanism is poorly understood. While it is expected that inactivation of p53 by E6 should lead to a reduction in apoptosis, E6 also sensitizes cells to apoptosis under some experimental conditions. In the present study, we demonstrated that expression of E6 in human keratinocytes rendered sensitization to chemotherapeutic agents. The cell death was shown to be by apoptosis involving caspase activation and the mitochondria pathway. To explore mechanisms involved in sensitization of E6 expressing cells to apoptosis, we used a proteomic approach to identify proteins differentially expressed in E6 expressing and control keratinocytes. Among nearly a thousand proteins examined, Cdc2 was demonstrated to be the most dramatically up-regulated protein in E6 expressing cells. p53 degradation appears to be important for the up-regulation of Cdc2 by E6. Using genetic, pharmacologic, and siRNA strategies, a role for Cdc2 in E6 expression-conferred apoptosis was demonstrated. Thus these results have important therapeutic implications in enhancing the efficacy of chemotherapy.
Keywords: HPV, E6, p53, apoptosis, Cdc2
Introduction
“High-risk” types of human papillomavirus (HPV) infect the anogenital tract and are strongly associated with the development of cervical carcinoma (for review, see 1). The transforming properties of high-risk HPVs primarily reside in the E6 and E7 genes 2; 3; 4; 5. The ability of the E6 protein to associate with the cellular tumor suppressor p53 has been suggested as a mechanism by which HPV induces tumors. The oncogenic activities of papillomavirus E6 have been reflected in many biological assays 6.
Apoptosis is a genetically programmed process of cellular destruction that is indispensable for the normal development and homeostasis of multi-cellular organisms 7. Apoptosis serves to eliminate damaged, aberrantly growing, or virally infected cells that could be a threat to the survival of an organism. Defects in apoptosis are thought to play a role in both the development of malignancy and resistance to chemotherapy 8; 9. While reduced apoptosis is implicated in cervical carcinoma, increased apoptosis has been observed in cervical intraepithelial neoplasia 10; 11. Although anti-apoptotic activities of papillomavirus E6 have been described in cultured cells, E6 could also induce or sensitize cells to apoptosis 6. The outcome of E6 modulation on apoptosis depends on experimental conditions. While the anti-apoptotic function of E6 can be attributed in part to its ability to degrade p53, little is known about how E6 sensitizes cells to apoptosis. We have recently demonstrated that down-regulation of p21 contributes to E6-induced apoptosis 12. How p21 inhibits apoptosis and what other targets, including those downstream from p21, are involved in E6-mediated apoptosis remain to be determined.
Activation of p53 by DNA damage induces either cell cycle arrest or apoptosis 13. The cytostatic effect of p53 is mediated by transcriptional activation of the cyclin-dependent kinase inhibitor p21 and 14-3-3 sigma, whereas the apoptotic effect is mediated in part by activation of proapoptotic gene products, including BAX 14; 15; 16; 17. Although inactivation of the p53 pathway is believed to be responsible for a reduction of apoptosis in cancers, some p53-deficient cells, including human keratinocytes expressing HPV E6, are more sensitive to certain DNA-damaging and chemotherapeutic agents (18; 19; 20; 21). The mechanism by which p53-defective cells are sensitized to apoptosis is not clear. Since p53 mutation, deletion, or down-regulation is common in cancers, it is important to develop strategies to induce apoptosis in the absence of wild-type p53.
The cyclin-dependent kinase 1 (Cdk1), also known as Cdc2 or p34Cdc2, interacts with cyclin B1 to form an active hetero-dimer, whose activity determines the timing of entry into mitosis 22. Activation of Cdc2 at the G2/M transition is precisely regulated through accumulation of cyclin B and a series of phosphorylation and dephosphorylation events of Cdc2 23. The DNA damage-responsive G2 checkpoint is able to delay mitosis by preventing the activation of Cdc2 24. p53 negatively regulates Cdc2 activity, inhibits the transcription of Cdc2/cyclin B1, and plays a role in the G2/M checkpoint (reviewed in 25). Several transcriptional targets of p53, including p21, can inhibit Cdc2 (26 and references therein). Cdc2 activation induces apoptosis in a variety of cell types (27; 28 and references therein), although several studies also showed that inactivation of Cdc2 increased the level of apoptosis 29; 30. In the present study, we provide evidence that activation of Cdc2 plays an important role in regulating apoptosis in E6 expressing keratinocytes.
Results
Expression of HPV E6 sensitizes cells to therapeutic agents
To explore the mechanism by which E6 sensitizes cells to apoptosis, we have established a cell culture system based on the expression of E6 from HPV type 16 (HPV-16), the most common high-risk HPV type, in spontaneously immortalized human foreskin keratinocytes (NIKS cells) 31. NIKS cells exhibit many characteristics of early-passage human keratinocytes, including the ability to stratify, differentiate, and sustain the HPV life cycle 31; 32. The use of immortalized cells for HPV oncogene studies can avoid comparisons being made between senescing (vector) and proliferating (E6) cells, when the experimental goal is to explore the mechanism of apoptosis. In addition, while primary human keratinocytes (PHKs) do not proliferate efficiently, NIKS cells grow relatively well in culture.
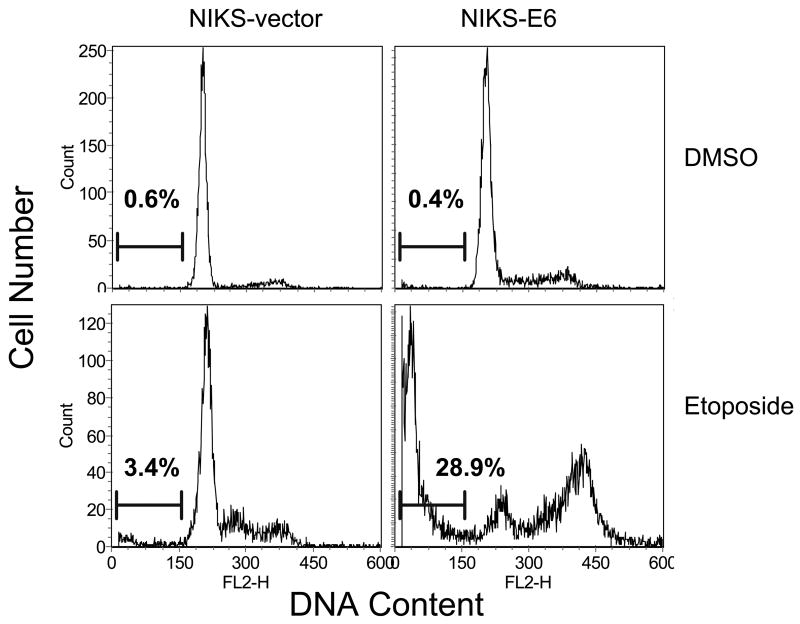
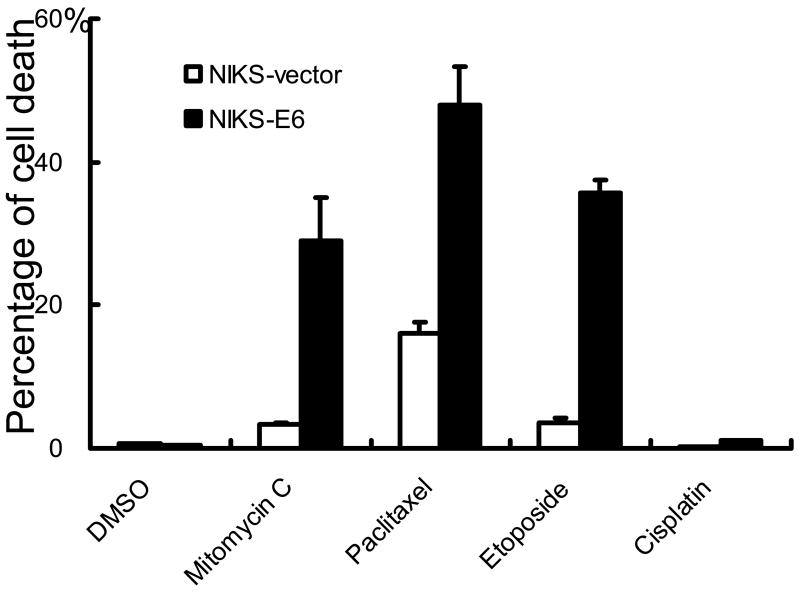
Accordingly, NIKS cells were infected with amphotrophic retroviruses containing HPV-16 E6 or the pBabe Puro vector. Populations of infected cells were selected and pooled. E6 expression was confirmed by real-time PCR (not shown). To evaluate the effect of E6 expression on cell death, NIKS-E6 cells were treated with multiple agents and cell death was determined by assessment of DNA fragmentation by flow cytometric analysis. As shown in Fig. 1A and 1B, a significant increase of cells with DNA content less than G1 (sub-G1), which represents cells with fragmented DNA and is one of the characteristics seen in cells undergoing apoptosis, was observed in NIKS-E6 but not vector cells after treatment with mitomycin C (MMC), Taxol (paclitaxel), and etoposide. In contrast, E6 expression did not significantly affect cell death in response to cisplatin (Fig. 1B), growth factor deprivation, tumor necrosis factor alpha, Fas ligand, and calcium (not shown). Since E6 is almost always expressed together with E7 in cervical cancers, we have examined the combined effect of E6 and E7 on apoptosis. As shown in Fig. S1, in response to etoposide, E6/E7 also sensitized cells to apoptosis and the combined effect of E6 and E7 is significantly more than E6 alone. We also determined cell death by analysis of MTT conversion 33. Consistent with what was observed in the DNA fragmentation analysis, NIKS-E6 cells showed significant increase in cell death in response to etoposide, MMC, and paclitaxel (Fig. S2A). Upon detailed examination, the effect of etoposide on cell death in NIKS-E6 cells was found to be dose-dependent and time-dependent (Fig. S2B and S2C).
Figure 1.
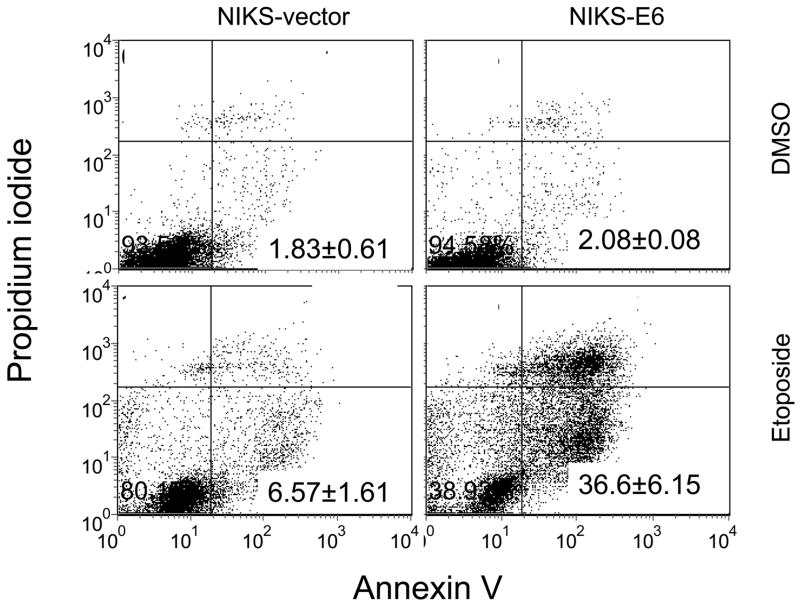
Expression of E6 sensitizes human keratinocytes to therapeutic agents. NIKS-E6 and vector control cells were treated with DMSO, MMC (5 μg/ml), paclitaxel (10 μM), etoposide (50 μM), or cisplatin (100 μM) for 48 hours. Cells were fixed, stained with propidium iodide (PI) or PI plus Annexin V-FITC, and analyzed by flow cytometry. Representative histograms of PI-stained, etoposide- or DMSO-treated cells from three experiments are shown in A. Quantification of sub-G1 populaiton of PI-stained cells is summarized in B. The results of a representative experiment (of 3) of etoposide/DMSO-treated, Annexin V-FITC/PI-stained cells are shown in C.
The cellular sensitization with E6 expression is not just restricted to keratinocytes. Expression of HPV E6 in hTERT-expressing human retinal pigment epithelium cells (RPE1) also increased cell death upon etoposide treatment (Fig. S3A). We also examined the sensitivity of cervical cancer cell lines CaSki and SiHa to therapeutic drugs. These cells are HPV-positive and therefore express E6 as well as E7 34. As shown in Fig. S3B, cervical cancer cells are highly sensitive to paclitaxel but are relatively resistant to cisplatin, a result consistent with what was observed in NIKS cells expressing HPV E6 (Fig. 1B).
Taken together, our results demonstrate that expression of HPV E6 sensitizes cells to therapeutic drugs. Since E6 degrades p53, the cell death observed does not appear to depend on the p53 pathway but rather related to p53 loss. Notably, in E6 expressing cells where p53 is down-regulated, chemotherapeutic drugs induce a G2/M arrest (Fig. 1A and S3A). Abrogation of G1 checkpoint by E6 has been documented and explains why cells do not arrest at the G1/S transition upon drug treatment. We consider the G2/M arrest could be induced by mechanisms including a p53-independent response to DNA damage, incomplete loss of p53, and the decatenation checkpoint that was shown to be independent of p53 35.
Apoptosis via caspase activation and the mitochondrial pathway is a potential mechanism of cell death in E6 expressing keratinocytes
To confirm that expression of E6 sensitized cells to therapeutic agents by inducing apoptosis, etoposide-treated NIKS-E6 cells were analyzed by Annexin V staining. This assay detects translocation of phosphatidylserine (PS) from the inner side of the plasma membrane to the outer layer, which occurs during early stages of apoptosis or necrosis. The latter can be excluded by simultaneous stain of the cells with PI. Consistent with the results from the DNA fragmentation analysis, increased Annexin V staining of the PI-negative population was found in NIKS-E6 cells after etoposide treatment (Fig. 1C).
Although NIKS and RPE1 cells contain the wild-type p53 sequence, some pathways leading to the activation of p53 may be disrupted. To alleviate this concern, we established PHKs expressing vector or HPV-16 E6 and their cellular sensitivity to etoposide was analyzed. As shown in Fig. S4, treatment with etoposide led to an increase in the steady-state level of p53 in PHKs expressing vector but not E6. These results demonstrate that the DNA damage pathway leading to p53 activation is functioning in PHKs and E6 is functionally active. Consistent with what was observed in NIKS and RPE1 cells, E6 expression in PHKs led to increased apoptosis upon etoposide treatment (data not shown).
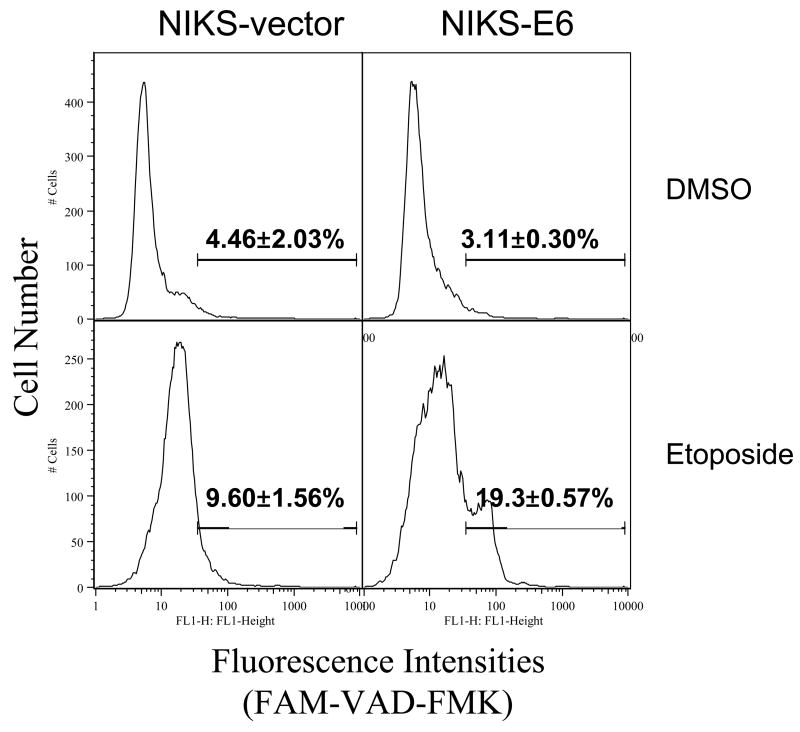
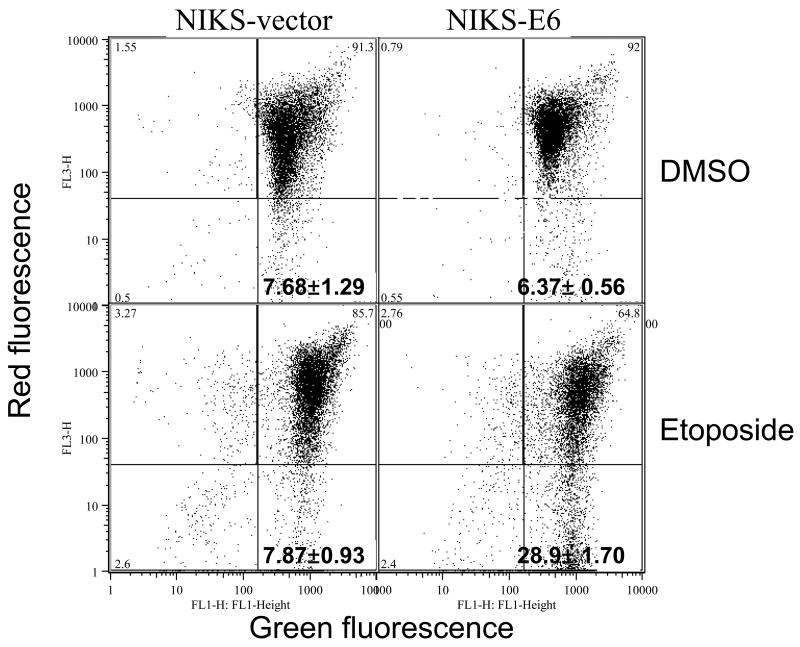
Caspase activation is usually associated with DNA fragmentation during apoptosis. To examine caspase activation, we labeled etoposide-treated NIKS-E6 cells with fluorescently labeled caspase inhibitor FAM-VAD-FMK, which specifically binds to active caspases. As shown in Fig. 2A, the population of NIKS-E6 cells with higher fluorescence intensity was significantly increased after etoposide treatment. These results demonstrate that caspases are activated during apoptosis in E6 expressing keratinocytes. Some apoptosis may occur via the mitochondrial pathway where the mitochondrial membrane potential (mtMP) collapses. To test this, we employed a cationic dye (JC-1) that forms red aggregates in normal mitochondria and remains in the cytoplasm as a green monomer upon collapse of the mtMP. Fig. 2B shows that treatment of NIKS-E6 cells with etoposide resulted in a decrease of the red aggregates of JC-1, indicating loss of mtMP.
Figure 2.
Activation of caspase and loss of mtMP are seen in E6 expressing cells undergoing apoptosis. E6 or vector control NIKS cells were treated with etoposide (50 μM) or DMSO for 48 hours. A, Cells were then collected and labeled with FAM-VAD-FMK for one hour. A representative experiment (of 2) is shown. Numbers in histograms correspond to percentage of caspase positive cells. B, Cells were labeled with JC-1 and analyzed on a flow cytometer. Dot plots are red fluorescence versus green fluorescence. A representative experiment (of 2) is shown.
Since loss of p53 has been implicated in mitotic catastrophe 36, a type of cell death that occurs during mitosis, we examined whether keratinocytes expressing E6 also undergo mitotic catastrophe upon DNA damage. We determined the number of NIKS-E6 cells undergoing mitosis upon etoposide-treatment. Since there are some similarities between cells in mitosis and cells undergoing apoptosis upon DAPI staining, we measured histone H3 phosphorylation, which occurs from prophase to late telophase 37. Our results demonstrated that few NIKS-E6 cells progressed into mitosis but rather first arrested at G2 before undergoing apoptosis after etoposide treatment, implying that these cells were not dying in mitosis (Fig. S5A). To rule out the possibility that lack of mitotic NIKS-E6 cells seen upon etoposide treatment is not a result of rapid exit of mitosis, nocodazole was added 5 hours after addition of etoposide to trap cells that may have progressed into mitosis. As shown in Fig. S5B, very few cells were detected in mitosis when treated with etoposide together with nocodazole whereas a significant number of NIKS-E6 cells were arrested in mitosis when treated with nocodazole alone (Fig. S5C). These results indicate that mitotic catastrophe is unlikely to be a major mechanism of cell death in NIKS-E6 cells upon treatment with a DNA damaging agent. As showed in Figure S5D, upon etoposide treatment, cells first arrest at G2 before undergoing apoptosis, suggesting that cells are dying at G2 phase.
Multiple proteins are differentially expressed in E6 expressing keratinocytes undergoing apoptosis
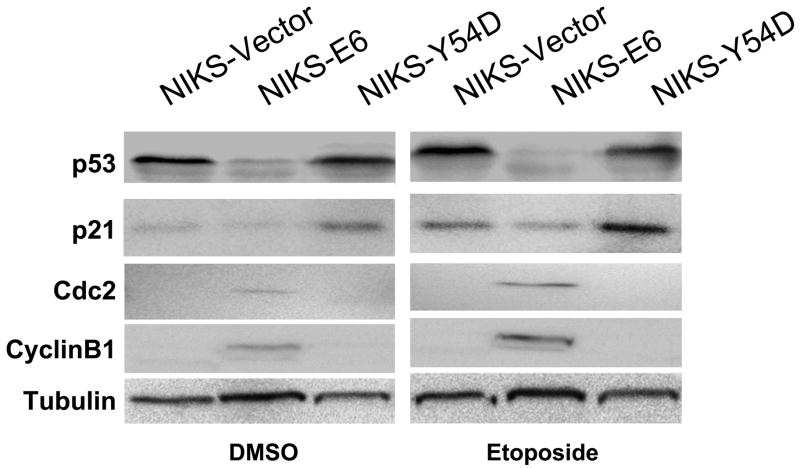
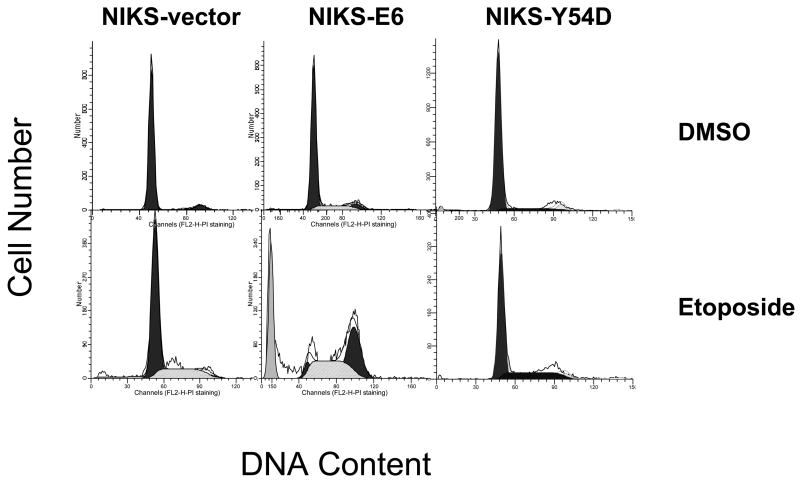
To identify proteins that may play a role in sensitizing cells to apoptosis in E6 expressing keratinocytes, we employed the PowerBlot Western Array Screening Service (BD Biosciences Pharmingen), which is a high-throughput immunoblotting technique that uses hundreds of antibodies targeted to a wide-range of relatively well-characterized cellular proteins. As a control, NIKS cells expressing HPV-16 E6 mutant Y54D (Tyr54 to Asp) were established. The E6 mutant Y54D is defective for p53 degradation and p53-mediated G1 arrest in response to DNA damage, yet is competent for immortalization of human mammary epithelial cells 12; 38. Therefore, we considered Y54D to be a better control than the vector. Y54D expression in NIKS cells was confirmed by real-time PCR (not shown). Western blot analysis confirmed that the steady-state levels of p53 and p21 were not significantly reduced in Y54D expressing NIKS cells (Fig. 3A). As expected, no significant apoptosis was observed in the NIKS-Y54D cells in response to etoposide treatment (Fig. 3C). Accordingly, protein expression between NIKS-E6 and NIKS-Y54D was compared. Of 921 monoclonal antibodies used, 802 generated valid signal. Among them, 90 protein bands exhibited a statistically significant (P<0.05) difference between NIKS-E6 and NIKS-Y54D cells treated with etoposide (Table S1). Proteins with the most dramatic changes (>2 folds) and statistically significant differences in steady-state levels are shown in Table 1. These include eight down-regulated proteins and 27 up-regulated proteins in NIKS-E6 cells compared to NIKS-Y54D cells.
Figure 3.
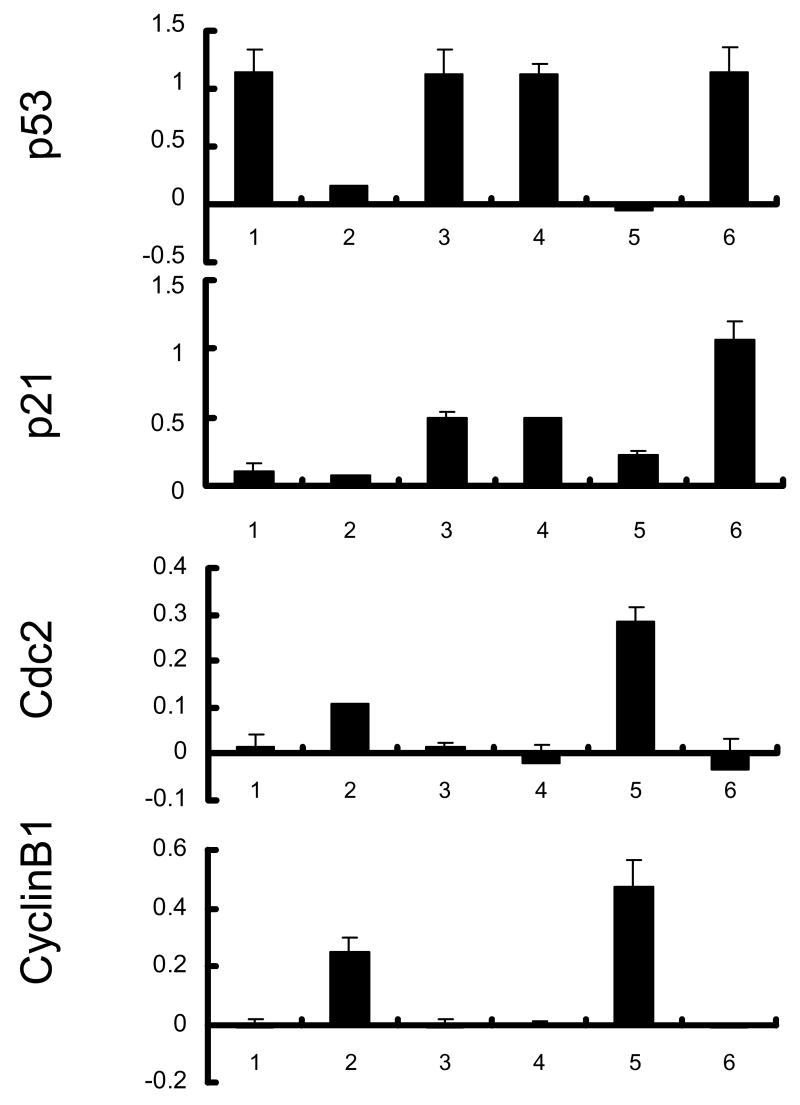
Cdc2 activity correlates with etoposide-induced apoptosis in E6 expressing cells. NIKS cells expressing E6 or E6 mutant Y54D were treated with etoposide (50 μM) or DMSO for 48 hours. A, Total protein extracts were resolved on SDS-PAGE and blotted with antibodies against p53, p21, Cdc2, and cyclin B1. β-tubulin blot was used as a loading control. B, Images of the blot in A were scanned and normalized against Tubulin loading control. Data represent the mean and standard deviation from two experiments. Columns 1 and 4, NIKS-Vector cells; 2 and 5, NIKS-E6 cells; 3 and 6, NIKS-Y54D cells; 1–3, DMSO-treated; 4–6 Etoposide-treated. C, NIKS cells expressing E6 or E6 mutant Y54D were fixed, stained with PI, and quantified for sub-G1 populations by flow cytometry. The program ModFit was used to analyze the cell cycle profile. A representative histogram 3 experiments is shown.
Table 1.
Differentially expressed proteins in HPV-16 E6 expressing keratinocytes
| Name | LocusLink ID | SwissProt ID | Fold changes (E6/Y54D) |
|---|---|---|---|
| Cdk1/Cdc2 | 983 | P06493 | * |
| MCM6 | 4175 | Q14566 | * |
| Beclin | 8678 | Q14457 | 3.43 |
| FEN-1 | 2237 | P39749 | 3.33 |
| Flotillin-1 | 14251 | O08917 | 3.29 |
| FADD | 8772 | Q13158 | 3.00 |
| PI3-Kinase C2 β | 5287 | O00750 | 2.96 |
| Cyclin A | 890 | P20248 | 2.66 |
| Jun | 16476 | P05627 | 2.64 |
| DNA Polymerase ε cat. | 5426 | Q07864 | 2.64 |
| Nestin | 10763 | P21263 | 2.64 |
| Karyopherin β 2 | 3838 | P52292 | 2.63 |
| PBK | 55872 | Q9NPD9 | 2.60 |
| GS15 | 54400 | O35152 | 2.58 |
| Cip1/WAF1 | 1026 | P38936 | 2.51 |
| Dystrobrevin | 13527 | Q9D2N4 | 2.50 |
| V-1/myotrophin | 94351 | P80144 | 2.46 |
| Glutamine Synthetase | 2752 | P15104 | 2.45 |
| Ron alpha | 4486 | Q04912 | 2.36 |
| Glucocorticoid R | 2908 | P04150 | 2.24 |
| Eg5 | 3832 | P52732 | 2.24 |
| p38 delta/SAPK4 | 5603 | O15264 | 2.16 |
| Na,K ATPase β 2 | 482 | P14415 | 2.14 |
| Plakophilin 2a | 5318 | Q99959 | 2.12 |
| FBP | 8880 | Q96AE4 | 2.10 |
| DRBP76 | 3609 | Q12906 | 2.01 |
| p160 | 10514 | O35851 | 0.50 |
| basic FGF-21kD | 2246 | P09038 | 0.49 |
| p115 | 56042 | P41542 | 0.48 |
| p53 | 7157 | P13481 | 0.45 |
| Vesl-1L | 65137 | O88800 | 0.45 |
| L-Caldesmon | 800 | Q05682 | 0.41 |
| p140mDia | 13367 | O08808 | 0.37 |
| hILP/XIAP | 331 | P98170 | 0.27 |
Proteins with more than 2-fold changes and statistically significant differences on the steady-state level between NIKS-E6 cells and NIKS-Y54D cells.
, represents presence of a protein in NIKS-E6 cells versus an absence in NIKS-Y54D cells of a protein.
As expected, a reduced level of p53 was detected in NIKS-E6 cells. However, the p21 level was not consistent with what we observed by Western blot (Fig. 3A). This might be attributed to the low signal of the p21 blot. Interestingly, Cdc2 had the most dramatically changed protein level. In addition and as expected, the level of multiple apoptosis-related proteins including beclin, FEN-1, FADD, Jun, glucocorticoid receptor (GR), FBP, and p38δ/SAPK4 was also significantly increased in E6-expressing cells. Among these proteins, beclin is an autophagy protein that can augment apoptosis 39, the flap endonuclease FEN-1 plays a role in DNA degradation during apoptosis 40, FADD is a death domain containing signaling adaptor protein during death receptor-induced apoptosis 41, activation of Jun has been implicated in apoptosis 42, glucocorticosteroid hormones induce apoptosis in several cell types 43, the FUSE-binding protein FBP is required for c-myc expression 44, and p38 d/SAPK4 plays a role in PKC delta-activated apoptosis 45. On the other hand, hILP/XIAP, which is related to the baculovirus iap gene and encodes an apoptosis inhibitor 46, was reduced in E6-expressing cells.
Since Cdc2 is the most dramatically changed protein in E6-expressing cells that undergo apoptosis and its activity has previously been implicated in inducing apoptosis in many experimental settings, we have focused on exploring the role of Cdc2 in apoptosis of E6 expressing cells. The increased protein level of Cdc2 in NIKS-E6 cells was confirmed by Western blot (Fig. 3A). In addition, the steady-state level of the Cdc2 partner cyclin B1 was also increased in NIKS-E6 cells (Fig. 3A). As increases in steady-state levels of Cdc2 and cyclin B1 does not necessarily lead to an increase in activity, we examined the kinase activity of Cdc2 in E6 expressing cells. The Cdc2-associated kinase activity was significantly increased in NIKS-E6 cells compared with the vector control cells (data not shown). The elevation in Cdc2/cyclin B1 expression and associated kinase activity in E6 expressing cells suggest that they play a role in sensitizing cells to apoptosis in these cells.
Role of the Cdc2/cyclin B1 complex in sensitization of E6 expressing cells to apoptosis
To examine the role of Cdc2 in apoptosis of E6 expressing cells, we took a genetic approach by comparing the ability of an E6 mutant to activate Cdc2 and to induce apoptosis in response to a therapeutic agent. First, the levels and kinase activities of Cdc2/cyclin B in E6 mutant Y54D cells were determined. As shown in Fig. 3A and 3B, neither Cdc2 nor cyclin B levels went up in the NIKS-Y54D cells. Consistent with these results, the Cdc2-associated kinase activity was low in NIKS-Y54D cells (data not shown). Since no significant apoptosis was observed in the NIKS-Y54D cells (Fig. 3C), a correlation between Cdc2 activity and apoptosis is implicated. As the E6 mutant Y54D is defective for p53 degradation, down-regulation of p53 correlates with up-regulation of Cdc2 by E6.
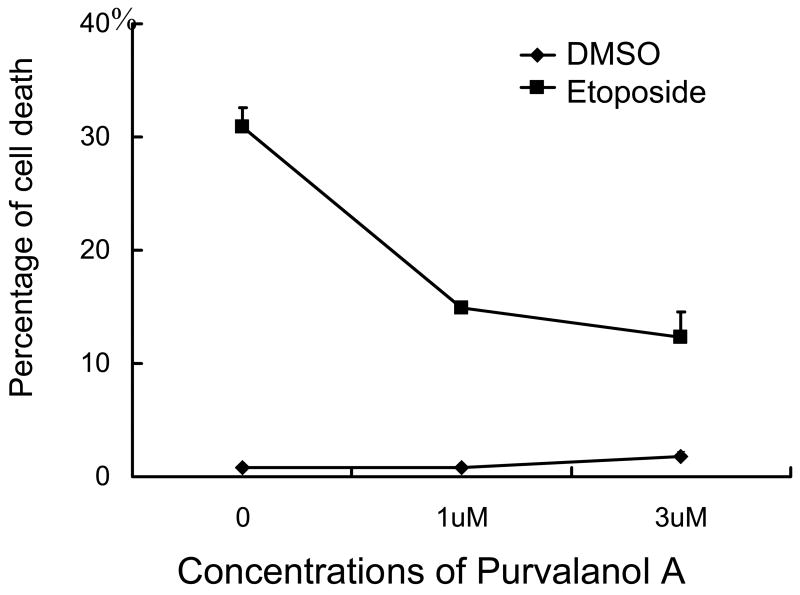
The role of Cdc2/cyclin B1 in sensitizing E6 expressing cells to apoptosis was then explored by a pharmacological approach using Purvalanol A, a potent Cdk inhibitor with a high selectivity for Cdc2 (In vitro IC50 = 4 nM for Cdc2/cyclin B, 35–70 nM for Cdk2, 850 nM for Cdk4, and virtually no effect on non-cdk protein kinases) 47. Addition of a low micro molar concentration of Purvalanol A to NIKS-E6 cells treated with etoposide significantly reduced apoptosis (Fig. 4A and S6A, P<0.05). Etoposide induces DNA damage, inhibits decatenation, and leads to G1/G2 arrests. A recent study demonstrates that Cdc2 also plays a role in G1/S transition 48, which is the major checkpoint in the cell. Inhibition of Cdc2 will arrest E6 cells at G1 and reduce the number of cells moving to G2 (Fig. S6A). These results support the notion that Cdc2 activity is required for induction of apoptosis in E6 expressing cells.
Figure 4.
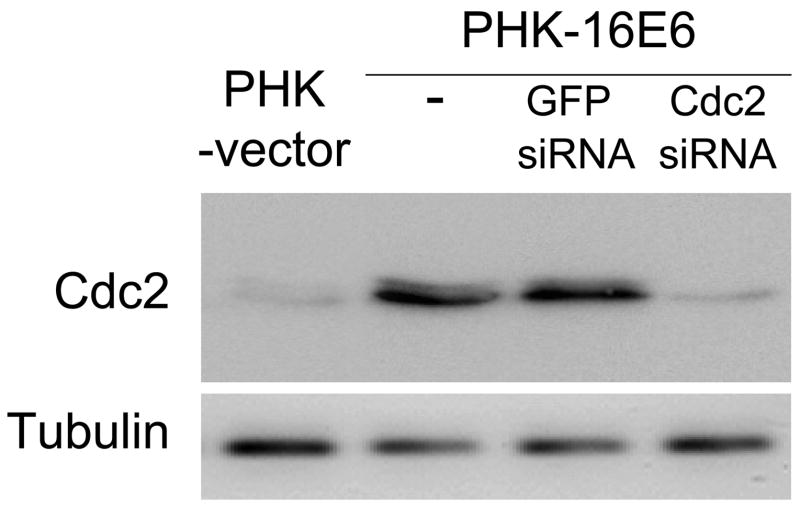
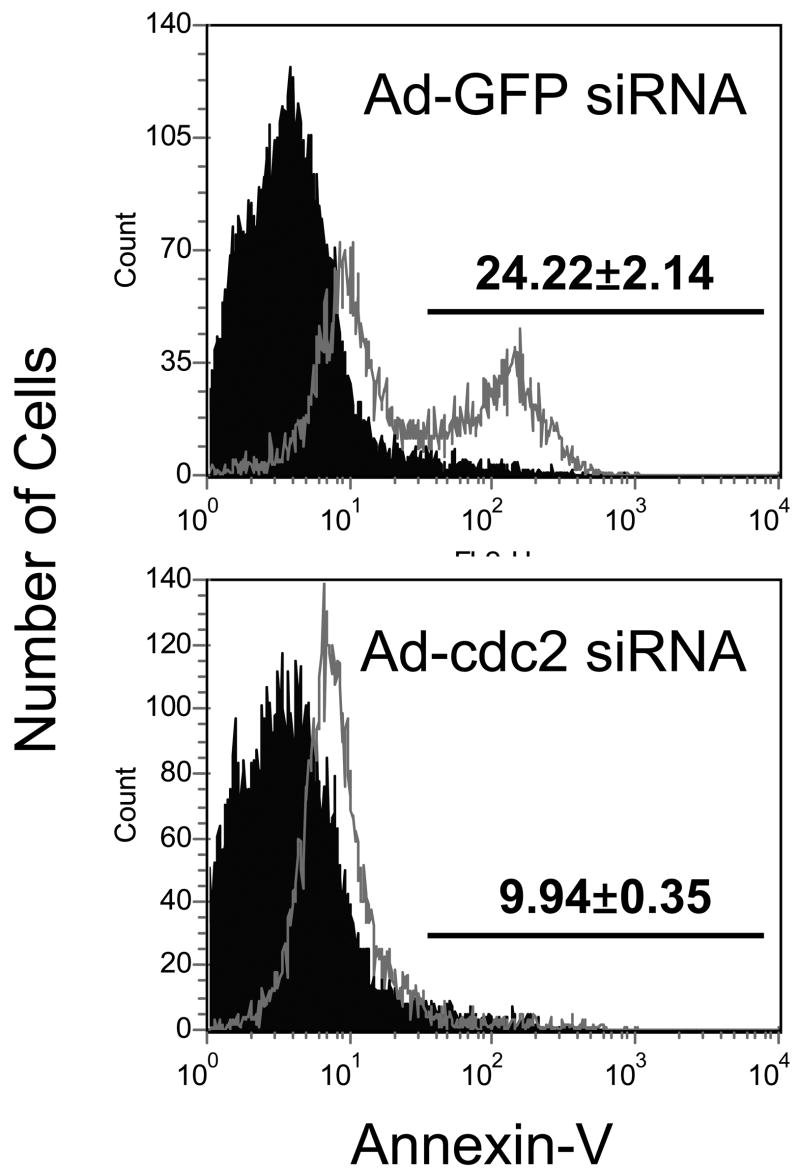
Cdc2 plays an important role in sensitization to etoposide-induced apoptosis in E6 expressing cells. A, Pharmacologic inhibition of Cdc2 activity reduces apoptosis in E6 expressing cells. NIKS-E6 cells were incubated with either DMSO or etoposide (50 μM) in the presence of Purvalanol A (Calbiochem, 1 μM or 3 μM) for 48 hours. DNA content was determined by flow cytometry after PI staining. The data represent an average of two experiments, each done in duplicate. Error bars represent standard deviations of the means. B, C, and D, Reduction of cdc2 expression by RNAi leads to a decrease of etoposide-induced apoptosis in E6-expressing cells. PHKs expressing E6 were infected with mock adenovirus or adenovirus expressing siRNAs targeting Cdc2 (297) or GFP at a titer of 5 × 106 IFU/ml. Forty-eight hours later, total protein extracts were subjected to Western blot using the Cdc2 antibody (B). In another set of experiments, eight hours after adenovirus infection with GFP siRNA or Cdc2 siRNA, PHKs were treated with etoposide (10 μM) for 48 hours, stained with Annexin V-FITC and PI, and analyzed by flow cytometry. The results of a representative experiment (of 2) showing Annexin V-positive fractions from PI-negative populations are given (C). Dark peaks, DMSO-treated; white peaks, etoposide-treated.
To further elucidate the role of Cdc2 in apoptosis in E6 expressing cells, an RNA interference strategy was used. We designed four small interfering RNAs (siRNAs) targeting Cdc2. The ability of the siRNAs to inhibit the activity of Cdc2 was examined for induction of a G2 cell cycle arrest in U2OS cells. As shown in Fig. S6B, among the four siRNAs, three efficiently induced G2 arrest after transient transfection. The siRNA 297 has the most dramatic inhibitory effect and was thus used in subsequent experiments. To facilitate efficient cellular delivery of siRNA, we employed the adenovirus system. The recombinant adenoviruses encoding siRNA 297 and a GFP siRNA were constructed and used to infect PHKs expressing HPV E6. Consistent with the G2 arrest results, the steady-state level of Cdc2 but not β-tubulin was significantly reduced in PHKs expressing E6 after infection with the adenovirus encoding siRNA 297 (Fig. 4B). In contrast, siRNA targeting GFP did not affect the steady-state level of Cdc2. We then examined the effect of siRNA 297 on etoposide-induced apoptosis in PHKs expressing E6. Significantly, etoposide-induced apoptosis was greatly reduced (to more than 50%) in PHKs expressing E6 infected with adenovirus encoding siRNA 297 but not siRNA targeting GFP (Fig. 4C). Taken together, these results indicate that Cdc2 is important for inducing apoptosis in E6 expressing cells in response to etoposide treatment.
Discussion
In the present study, we demonstrated that expression of HPV E6 rendered human keratinocytes to be sensitized to DNA damaging and chemotherapeutic agents. We further showed that the cell death was apoptosis involving caspase activation and the mitochondria pathway. A role for Cdc2 in E6-mediated apoptosis was demonstrated. p53 degradation appears to be important for the up-regulation of Cdc2 by E6. Although activation of Cdc2 by E6 has been shown previously 49 and Cdc2-induced apoptosis has been implicated, this is the first study to demonstrate a critical role of Cdc2 in E6-mediated apoptosis. These results have important therapeutic implications in cancer therapy.
It is well established that activation of p53 induces apoptosis in many systems. However, loss of p53 also sensitizes tumor cells to certain therapeutic agents in some experimental conditions 20. The mechanism by which disruption of p53, through expression of HPV E6 in particular, enhances apoptosis is not well understood. p53 can induce both cell-cycle arrest or apoptosis, depending on cell types and reagents used to treat the cells. It remains to be shown what determines the cell type specificity and how individual reagent selectively induces p53-dependent apoptosis rather than cell cycle arrest. We speculate that down-regulation of p53-induced apoptosis will more likely occur in the types of cells where wild-type p53 would readily induce cell-cycle arrest rather than apoptosis in response to DNA damage.
The role of Cdc2 in apoptosis remains a conundrum. While our results, along with many other studies, demonstrate that Cdc2 activation induces apoptosis, several other studies showed that inactivation of Cdc2 leads to increased apoptosis (50 and references therein). The discrepancy regarding the contribution of Cdc2 to cell death could be due to cell-type specificity and/or experimental conditions. It remains a possibility that the observed Cdc2 activation is a consequence rather than a cause of apoptosis. However, this is unlikely to be the case in our system where activity of Cdc2 has been elevated before treatment with DNA damaging agents.
Notably, in etoposide-treated E6 expressing cells, although Cdc2 remains active, cells arrested at S/G2 stage (Fig. 1A, 3C, S3A, S5, and S6A). These results suggest that Cdc2 activation is not sufficient for abrogating the G2 checkpoint in E6 expressing cells. These results are also consistent with a previous observation 51. In addition, the decatenation checkpoint triggered by etoposide can also arrest cells at G2/M that is independent of the Cdc2 activity. We hypothesis that Cdc2 mainly promotes cell death at the G2. Inhibition of Cdc2 activity in E6 expressing cells that were treated with etoposide increased cells in the G1 stage and decreased cells in G2 and sub-G1 (Figure S6A). The G1 arrest induced by Cdc2 inhibition is consistent with a role for Cdc2 in G1/S transition 48.
In this study, we have focused our effort on exploring the role of Cdc2/cyclin B in apoptosis in E6 expressing human keratinocytes. We understand that activation of Cdc2/cyclin B may not be the only mechanism by which E6 mediates apoptosis. Multiple proteins among those that are differentially expressed in E6 expressing cells undergoing apoptosis have been identified. Cyclin A, which is up-regulated in E6 expressing cells, can form a complex with Cdc2 and may contribute to E6-mediated apoptosis. FADD is the principal mediator of death receptor signaling in mammalian cells. However, FADD is involved in the extrinsic pathway while the apoptosis described here is through the intrinsic pathway. Up-regulation of FADD could simply be a consequence rather than a mechanism underlying E6-mediated apoptosis. Notably, the c-Jun N-terminal kinase, which phosphorylates c-Jun and enhances its transcriptional activity, has been demonstrated to have a role in p53-independent apoptosis 52; 53. Future studies should be designed to explore the role of these proteins in apoptosis in E6 expressing cells. Although we have demonstrated that apoptosis is a mechanism of cell death in E6 expressing human keratinocytes in response to therapeutic agents, we can not rule out the possibility that non-apoptotic programmed cell death, such as necrosis or autophagy, may also contribute to cell death in this process. It remains to be determined what the non-apoptotic mechanism is by which E6 expressing cells are dying in response to therapeutic agents.
Our studies have important therapeutic implications. A desirable approach to treat a tumor is to selectively induce cancer cells to die by apoptosis. Some cancer therapies are rationalized according to the status of p53. However, mutation of the p53 gene is one of the most frequently reported genetic defects in human cancers. In case of cervical cancer, p53 is degraded in the presence of HPV E6. Understanding the mechanism of cellular sensitization to apoptosis in E6 expressing cells is important for the development of novel approaches for treating cancers. E6 expression in cancer cells can be explored to enhance the efficacy of therapeutic agents. On the other hand, our results call into caution cancer gene therapy that would restore wild-type p53 function.
Experimental procedures
Cell culture
NIKS cells are spontaneously immortalized human keratinocytes 31 originally available from ATCC (12191). NIKS cells were maintained on MMC-treated J2-3T3 feeder cells in F medium composed of 3 parts Ham’s F12 medium to 1 part Dulbecco’s modified Eagle’s medium (DMEM) with supplements as previously described 31. The PHKs were prepared from neonatal foreskins obtained from Xijing Hospital and maintained in Keratinocyte-Serum Free Medium (K-SFM, Invitrogen) supplemented with 5 ng/ml epidermal growth factor and 25 μg/ml bovine pituitary extract. To establish stable E6-expressing cell lines, plasmids encoding HPV-16 E6 and E6 mutant Y54D in the pBabe Puro vector were transfected into the amphotropic Phoenix™ retrovirus packaging cell line 54 by liposome-mediated transfection (FuGENE 6, Roche) as described previously 55. Viruses were collected 48h after transfection and titrated on NIKS cells to determine the puromycin-resistant colony forming units (cfu). NIKS cells or PHKs were then infected with retroviruses containing approximately equal cfu. After puromycin selection, populations of infected cells were pooled and used for subsequent experiments. To avoid the possibility of chromosomal instability due to the expression of HPV-16 E6, all experiments were performed using cells within 10 passages. The human osteosarcoma cell line U2OS and HPV-positive cervical cancer cell lines CaSki and SiHa cells were cultured in DMEM plus 10% fetal calf serum. RPE1 cells were originally obtained from Clontech and were cultured in medium composed of 1 part Ham’s F12 medium to 1 part DMEM with 10% fetal calf serum
Flow cytometry
For cell death and cell cycle analysis, cells were fixed in 70% ethanol overnight. Keratinocytes were treated by etoposide (Sigma, 50 μM), MMC (Sigma, 5 μg/ml), or paclitaxel (ICN Biomedicals, 10 μM) for 48h before analysis unless otherwise specified. After re-suspension in PBS, cells were incubated in staining solution containing PI (50 μg/ml, Sigma) or TO-PRO-3 (1 μM, Molecular Probes) supplemented with 100 μg/ml RNase A (Sigma). Samples were analyzed with a BD LSR flow cytometer. The cell cycle profiles were analyzed using FCS Express v3.0 (De Novo Software).
For Annexin V staining, cells (both floating and adherent) treated with therapeutic agents were harvested and pelleted, stained with PI and Phycoerythrin-conjugated Annexin-V. For active caspase analysis, treated cells were labeled with FAM-VAD-FMK (Immunochemistry Technologies, LLC). For measuring mtMP, the APO LOGIX JC-1 Assay Kit from Cell Technology was used. A dot plot of red fluorescence (FL2) versus green fluorescence (FL1) resolved live cells with intact mtMP from apoptotic and dead cells with lost mtMP.
Immunoblotting and kinase assay
Protein extraction was performed in lysis buffer A (10 mM Tris-HCl, pH 7.4, 1% sodium deoxycholate, 150 mM sodium chloride, 1.0 mM sodium ortho-vanadate). Equal amounts of protein from each cell lysate were separated in an SDS poly-acrylamide gel and transferred onto a PVDF membrane. Filters were blotted with antibodies against p53 (DO-1, BD bioscience), p21 (Cat. #610233, BD Bioscience), Cdc2 (Cat. #610037, BD Bioscience), cyclin B1 (Cat. #554176, BD Bioscience), and Tubulin (Cat. #T4026, Sigma). The kinase assay was performed essentially as described previously 56. Cells were lysed in lysis buffer B (50 mM HEPES, pH 7.5, 150 mM NaCl, 2.5 mM EGTA, 1 mM EDTA, 1 mM dithiothreitol, 10% glycerol, 0.1% Tween 20, and protease inhibitors mixture of aprotinin, leupeptin, and pepstatin A, and phenylmethylsulfonyl fluoride). 100 μg of total protein extract was immunoprecipitated with the anti-Cdc2 antibody and protein A-agarose (Amersham Biosciences). Assays were performed in the presence of 10 μCi of [γ-32P]ATP (3000 Ci/mmol, PerkinElmer Life Sciences), 20 μM cold ATP, and 2 μg of histone H1 (Sigma).
PowerBlot Screen
NIKS cells expressing wild-type HPV-16 E6 or mutant Y54D were treated with etoposide (50 μM) for 48 hours. Total cellular proteins were extracted using lysis buffer C (10 mM Tris-HCl, pH 7.4, 1.0 mM sodium ortho-vanadate, and 1% SDS). The primary screening and analysis were performed at BD Biosciences. Briefly, cell lysates were separated in multiple gradient SDS poly-acrylamide gels and transferred to Immobilon-P membranes (Millipore). Each membrane was divided into 40 channels and each channel was blotted with different sets of four to five mouse monoclonal antibodies that recognize proteins of distinct molecular weights. The membrane was then incubated with goat anti-mouse conjugated to Alexa680 fluorescent dye (Molecular Probes) and scanned using the Odyssey Infrared Imaging System (LI-COR). The bands were identified and normalized. The data from three independent samples were analyzed using a 3 × 3 matrix comparison method.
Recombinant adenovirus construction, purification, and titration
The AdEasy adenovirus system and the p53 plasmid were gifts from Dr. B. Vogelstein (John Hopkins Oncology Center, Baltimore, MD). Four Cdc2 siRNAs, including 202 (5′-ACTACAGGTCAAGTGGTAG); 297 (5′-GGAACTTCGTCATCCAAAT); 330 (5′-GGATGTGCTTATGCAGGAT); 905 (5′-ATGGCTTGGATTTGCTCTC, were designed according to the recommendations of Tuschl (The siRNA user guide: http://www.rockefeller.edu/labheads/tuschl/sirna.html, revised 2003). The recombinant adenoviruses encoding the p53 mutants, Cdc2 siRNA were produced as previously described 57. Briefly, p53 and H1 promoter-Cdc2 siRNA fragments were cloned into the shuttle vector pAdTrack-CMV and pAdTrack, respectively, linearized, and co-transformed into Escherichia coli BJ5183 cells along with the adenoviral backbone vector pAdEasy-1. Recombinants were selected for kanamycin resistance and confirmed by restriction endonuclease analyses. Finally, a linearized recombinant plasmid was transfected into an adenovirus packaging cell line, HEK293 cells. An adenovirus containing only EGFP was used as a control. In the viral infection experiments, adenoviruses were added at a titer of 5 × 106 infectious units (IFU)/ml, to the culture medium 8 hours before etoposide treatment.
Statistical Analysis
Data were expressed as mean ± S.E. Differences between means were assessed by Student’s t test. p = 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. B. Vogelstein for plasmids, L.X. Zhang for help with the keratinocytes preparation, Dr. Elliot Androphy for advice, Susan Heilman for critical reading and editing of the manuscript, Dr. Jianyuan Luo for helpful discussion and critical reading of the manuscript, and Dr. Junying Yuan for comments. This work was supported by grants to Z.G. Liu from National Scientific Foundation of China (grant number 30400016) and J.J.C. from the USA National Institutes of Health (R03AI060829).
Footnotes
Supplementary information is available at JMB’s website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.zur Hausen H. Papillomavirus infections-a major cause of human cancers. Biochimica et Biophysica Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]
- 2.Werness BA, Levine AJ, Howley PM. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science. 1990;248:76–79. doi: 10.1126/science.2157286. [DOI] [PubMed] [Google Scholar]
- 3.Dyson N, Howley PM, Munger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 4.Munger K, Werness BA, Dyson N, Phelps WC, Harlow E, Howley PM. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gage JR, Meyers C, Wettstein FO. The E7 proteins of the nononcogenic human papillomavirus type 6b (HPV-6b) and of the oncogenic HPV-16 differ in retinoblastoma protein binding and other properties. J Virol. 1990;64:723–730. doi: 10.1128/jvi.64.2.723-730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rapp L, Chen JJ. The papillomavirus E6 proteins. Biochim Biophys Acta. 1998;1378:F1–F19. doi: 10.1016/s0304-419x(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 7.Rich T, Watson CJ, Wyllie A. Apoptosis: the germs of death. Nat Cell Biol. 1999;1:E69–71. doi: 10.1038/11038. [DOI] [PubMed] [Google Scholar]
- 8.Teodoro JG, Branton PE. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodriguez-Nieto S, Zhivotovsky B. Role of alterations in the apoptotic machinery in sensitivity of cancer cells to treatment. Curr Pharm Des. 2006;12:4411–25. doi: 10.2174/138161206779010495. [DOI] [PubMed] [Google Scholar]
- 10.Duttagupta C, Basu J, Ray M, Romney SL. Apoptotic changes in cervical intraepithelial neoplasia. Gynecol Obstet Invest. 2001;52:38–42. doi: 10.1159/000052938. [DOI] [PubMed] [Google Scholar]
- 11.Chung TK, Cheung TH, Lo WK, Yim SF, Yu MY, Krajewski S, Reed JC, Wong YF. Expression of apoptotic regulators and their significance in cervical cancer. Cancer Lett. 2002;180:63–8. doi: 10.1016/s0304-3835(01)00842-4. [DOI] [PubMed] [Google Scholar]
- 12.Fan X, Liu Y, Chen JJ. Down-regulation of p21 contributes to apoptosis induced by HPV E6 in human mammary epithelial cells. Apoptosis. 2005;10:63–73. doi: 10.1007/s10495-005-6062-y. [DOI] [PubMed] [Google Scholar]
- 13.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 14.Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B. 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature. 1999;401:616–20. doi: 10.1038/44188. [DOI] [PubMed] [Google Scholar]
- 15.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 16.Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–82. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- 17.Nakano K, Vousden KH. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell. 2001;7:683–94. doi: 10.1016/s1097-2765(01)00214-3. [DOI] [PubMed] [Google Scholar]
- 18.Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW, Galloway DA. Loss of normal p53 function confers sensitization to taxol by increased G2/M arrest and apoptosis. Nat Med. 1996;2:72–79. doi: 10.1038/nm0196-72. [DOI] [PubMed] [Google Scholar]
- 19.Wang Q, Fan S, Eastman A, Worland PJ, Sausville EA, O’Connor PM. UCN-01: a potent abrogator of G2 checkpoint function in cancer cells with disrupted p53. J Natl Cancer Inst. 1996;88:956–65. doi: 10.1093/jnci/88.14.956. [DOI] [PubMed] [Google Scholar]
- 20.Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, Williams J, Lengauer C, Kinzler KW, Vogelstein B. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, McKalip A, Herman B. Human papillomavirus type 16 E6 and HPV-16 E6/E7 sensitize human keratinocytes to apoptosis induced by chemotherapeutic agents: roles of p53 and caspase activation. J Cell Biochem. 2000;78:334–49. [PubMed] [Google Scholar]
- 22.Pines J, Hunter T. p34cdc2: the S and M kinase? New Biol. 1990;2:389–401. [PubMed] [Google Scholar]
- 23.Nurse P. Checkpoint pathways come of age. Cell. 1997;91:865–867. doi: 10.1016/s0092-8674(00)80476-6. [DOI] [PubMed] [Google Scholar]
- 24.O’Connor PM, Ferris DK, Hoffmann I, Jackman J, Draetta G, Kohn KW. Role of the cdc25C phosphatase in G2 arrest induced by nitrogen mustard. Proc Natl Acad Sci U S A. 1994;91:9480–4. doi: 10.1073/pnas.91.20.9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor WR, Stark GR. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–15. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 26.Yu D, Jing T, Liu B, Yao J, Tan M, McDonnell TJ, Hung MC. Overexpression of ErbB2 blocks Taxol-induced apoptosis by upregulation of p21Cip1, which inhibits p34Cdc2 kinase. Mol Cell. 1998;2:581–91. doi: 10.1016/s1097-2765(00)80157-4. [DOI] [PubMed] [Google Scholar]
- 27.Shi L, Nishioka WK, Th’ng J, Bradbury EM, Litchfield DW, Greenberg AH. Premature p34cdc2 activation required for apoptosis. Science. 1994;263:1143–5. doi: 10.1126/science.8108732. [DOI] [PubMed] [Google Scholar]
- 28.Tan M, Jing T, Lan KH, Neal CL, Li P, Lee S, Fang D, Nagata Y, Liu J, Arlinghaus R, Hung MC, Yu D. Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell. 2002;9:993–1004. doi: 10.1016/s1097-2765(02)00510-5. [DOI] [PubMed] [Google Scholar]
- 29.Ongkeko W, Ferguson DJ, Harris AL, Norbury C. Inactivation of Cdc2 increases the level of apoptosis induced by DNA damage. J Cell Sci. 1995;108(Pt 8):2897–904. doi: 10.1242/jcs.108.8.2897. [DOI] [PubMed] [Google Scholar]
- 30.Itzhaki JE, Gilbert CS, Porter AC. Construction by gene targeting in human cells of a “conditional’ CDC2 mutant that rereplicates its DNA. Nat Genet. 1997;15:258–65. doi: 10.1038/ng0397-258. [DOI] [PubMed] [Google Scholar]
- 31.Allen-Hoffmann BL, Schlosser SJ, Ivarie CA, Sattler CA, Meisner LF, O’Connor SL. Normal growth and differentiation in a spontaneously immortalized near-diploid human keratinocyte cell line, NIKS. J Invest Dermatol. 2000;114:444–55. doi: 10.1046/j.1523-1747.2000.00869.x. [DOI] [PubMed] [Google Scholar]
- 32.Flores ER, Allen-Hoffmann BL, Lee D, Sattler CA, Lambert PF. Establishment of the human papillomavirus type 16 (HPV-16) life cycle in an immortalized human foreskin keratinocyte cell line. Virology. 1999;262:344–54. doi: 10.1006/viro.1999.9868. [DOI] [PubMed] [Google Scholar]
- 33.Hansen MB, Nielsen SE, Berg K. Re-examination and further development of a precise and rapid dye method for measuring cell growth/cell kill. J Immunol Methods. 1989;119:203–210. doi: 10.1016/0022-1759(89)90397-9. [DOI] [PubMed] [Google Scholar]
- 34.Hietanen S, Lain S, Krausz E, Blattner C, Lane DP. Activation of p53 in cervical carcinoma cells by small molecules. Proc Natl Acad Sci U S A. 2000;97:8501–6. doi: 10.1073/pnas.97.15.8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaufmann WK, Campbell CB, Simpson DA, Deming PB, Filatov L, Galloway DA, Zhao XJ, Creighton AM, Downes CS. Degradation of ATM-independent decatenation checkpoint function in human cells is secondary to inactivation of p53 and correlated with chromosomal destabilization. Cell Cycle. 2002;1:210–9. [PubMed] [Google Scholar]
- 36.Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat. 2001;4:303–13. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- 37.Juan G, Traganos F, Darzynkiewicz Z. Methods to identify mitotic cells by flow cytometry. Methods Cell Biol. 2001;63:343–54. doi: 10.1016/s0091-679x(01)63019-x. [DOI] [PubMed] [Google Scholar]
- 38.Liu Y, Chen JJ, Gao Q, Dalal S, Hong Y, Mansur CP, Band V, Androphy EJ. Multiple functions of human papillomavirus type16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Furuya D, Tsuji N, Yagihashi A, Watanabe N. Beclin 1 augmented cis-diamminedichloroplatinum induced apoptosis via enhancing caspase-9 activity. Exp Cell Res. 2005;307:26–40. doi: 10.1016/j.yexcr.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 40.Parrish JZ, Yang C, Shen B, Xue D. CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. Embo J. 2003;22:3451–60. doi: 10.1093/emboj/cdg320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–12. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 42.Estus S, Zaks WJ, Freeman RS, Gruda M, Bravo R, Johnson EM., Jr Altered gene expression in neurons during programmed cell death: identification of c-jun as necessary for neuronal apoptosis. J Cell Biol. 1994;127:1717–27. doi: 10.1083/jcb.127.6.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Distelhorst CW. Recent insights into the mechanism of glucocorticosteroid-induced apoptosis. Cell Death Differ. 2002;9:6–19. doi: 10.1038/sj.cdd.4400969. [DOI] [PubMed] [Google Scholar]
- 44.He L, Liu J, Collins I, Sanford S, O’Connell B, Benham CJ, Levens D. Loss of FBP function arrests cellular proliferation and extinguishes c-myc expression. EMBO J. 2000;19:1034–44. doi: 10.1093/emboj/19.5.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eckert RL, Efimova T, Balasubramanian S, Crish JF, Bone F, Dashti S. p38 Mitogen-activated protein kinases on the body surface--a function for p38 delta. J Invest Dermatol. 2003;120:823–8. doi: 10.1046/j.1523-1747.2003.12120.x. [DOI] [PubMed] [Google Scholar]
- 46.Duckett CS, Nava VE, Gedrich RW, Clem RJ, Van Dongen JL, Gilfillan MC, Shiels H, Hardwick JM, Thompson CB. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. Embo J. 1996;15:2685–94. [PMC free article] [PubMed] [Google Scholar]
- 47.Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, Morgan DO, Barnes G, LeClerc S, Meijer L, Kim SH, Lockhart DJ, Schultz PG. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors. Science. 1998;281:533–8. doi: 10.1126/science.281.5376.533. [DOI] [PubMed] [Google Scholar]
- 48.Aleem E, Kiyokawa H, Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat Cell Biol. 2005;7:831–6. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 49.Thompson DA, Belinsky G, Chang TH, Jones DL, Schlegel R, Munger K. The human papillomavirus-16 E6 oncoprotein decreases the vigilance of mitotic checkpoints. Oncogene. 1997;15:3025–3035. doi: 10.1038/sj.onc.1201495. [DOI] [PubMed] [Google Scholar]
- 50.O’Connor DS, Wall NR, Porter AC, Altieri DC. A p34(cdc2) survival checkpoint in cancer. Cancer Cell. 2002;2:43–54. doi: 10.1016/s1535-6108(02)00084-3. [DOI] [PubMed] [Google Scholar]
- 51.Passalaris TM, Benanti JA, Gewin L, Kiyono T, Galloway DA. The G(2) checkpoint is maintained by redundant pathways. Mol Cell Biol. 1999;19:5872–81. doi: 10.1128/mcb.19.9.5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C, Ma WY, Li J, Dong Z. Arsenic induces apoptosis through a c-Jun NH2-terminal kinase-dependent, p53-independent pathway. Cancer Res. 1999;59:3053–8. [PubMed] [Google Scholar]
- 53.Bai L, Yoon SO, King PD, Merchant JL. ZBP-89-induced apoptosis is p53-independent and requires JNK. Cell Death Differ. 2004;11:663–73. doi: 10.1038/sj.cdd.4401393. [DOI] [PubMed] [Google Scholar]
- 54.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci U S A. 1993;90:8392–6. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Z, Liu Y, Hong Y, Rapp L, Androphy EJ, Chen JJ. Bovine papillomavirus type 1 E6-induced sensitization to apoptosis is distinct from its transforming activity. Virology. 2002;295:230–7. doi: 10.1006/viro.2001.1351. [DOI] [PubMed] [Google Scholar]
- 56.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–83. doi: 10.1128/mcb.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He TC, Zhou S, da Costa LT, Yu J, Kinzler KW, Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–14. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.