Abstract
LINE-1 retrotransposons constitute one-fifth of human DNA and have helped shape our genome. A full-length L1 encodes a 40-kDa RNA-binding protein (ORF1p) and a 150-kDa protein (ORF2p) with endonuclease and reverse transcriptase activities. ORF1p is distinctive in forming large cytoplasmic foci, which we identified as cytoplasmic stress granules. A phylogenetically conserved central region of the protein is critical for wild-type localization and retrotransposition. Yeast two-hybrid screens revealed several RNA-binding proteins that coimmunoprecipitate with ORF1p and colocalize with ORF1p in foci. Two of these proteins, YB-1 and hnRNPA1, were previously reported in stress granules. We identified additional proteins associated with stress granules, including DNA-binding protein A, 9G8, and plasminogen activator inhibitor RNA-binding protein 1 (PAI-RBP1). PAI-RBP1 is a homolog of VIG, a part of the Drosophila melanogaster RNA-induced silencing complex (RISC). Other RISC components, including Ago2 and FMRP, also colocalize with PAI-RBP1 and ORF1p. We suggest that targeting ORF1p, and possibly the L1 RNP, to stress granules is a mechanism for controlling retrotransposition and its associated genetic and cellular damage.
During the course of human evolution, the LINE-1 retrotransposon has expanded to over 500,000 copies composing approximately 18% of the mass of the genome. In addition, the L1 retrotransposition machinery has been responsible for genomic insertion of over a million nonautonomous retroposons, including Alus and SVA elements, and approximately 8,000 processed pseudogenes (35, 86). L1 expansion has slowed dramatically, and only 19 recent disease-causing human insertions have been identified. Most L1 insertions are truncated, rearranged, or mutated and hence incapable of further retrotransposition. Nevertheless, there are 6,000 full-length human elements, among which roughly 80 to 100 are potentially capable of retrotransposition (7), but many more can be transcribed and translated.
The 6.0-kb full-length human L1 has a 906-nucleotide (nt) 5′ untranslated region (UTR) with an internal promoter, two open reading frames (ORF1 and ORF2) separated by a 63-bp intergenic spacer, and a 206-nt 3′ UTR which ends in a poly(A) signal and tail. L1 ORF2 encodes a 149-kDa protein (ORF2p) with endonuclease and reverse transcriptase activities. Unlike the ORF1 product, translation of ORF2p is suppressed, and its low copy number has made ORF2p difficult to find in most tissues and cell lines, even when it is overexpressed (16, 25).
ORF1 protein (ORF1p) is a nucleic acid-binding protein that lacks sequence similarity with any other known protein. Mutational analysis has shown it to be essential for retrotransposition (64), but its exact function remains unclear. The N-terminal one-third of both human and mouse ORF1p contains an α-helical coiled-coil domain, which is predicted to form a leucine zipper in humans (30). In mouse ORF1p, this region is necessary and sufficient for stable trimerization. The C-terminal-third of the protein is responsible for low sequence specificity and high-affinity binding of RNA and single-stranded DNA, and mouse ORF1p can act as an RNA chaperone (60-62). In sucrose gradients, both human and mouse ORF1 protein cofractionates with L1 RNA in a large ribonucleoprotein (RNP) complex, and reverse transcriptase activity linked to the presence of ORF2p in this RNP has been detected (29, 46, 58).
It is believed that L1s retrotranspose by a process termed target primed reverse transcription (56). According to this model, ORF2-encoded endonuclease nicks the bottom strand of target DNA and exposes a 3′-hydoxyl that primes reverse transcription of L1 RNA. Evidence from human insertions and cell culture indicates that L1 proteins have cis preference, binding their own encoding RNA (17, 82). To date, no non-L1 proteins have been identified as components of the L1 RNP. Almost certainly, however, other cellular factors bind the RNP and participate in the complex process of retrotransposition.
Here we focused on the subcellular distribution of human ORF1p and some of its associated non-L1 proteins. Both mouse and human ORF1 proteins are predominantly cytoplasmic when assayed by subcellular fractionation or immunofluorescence (29, 59). We previously reported that ORF2p, when overexpressed in a modified vaccinia virus Ankara/T7 RNA polymerase (MVA/T7RP) hybrid system, is also predominantly cytoplasmic and that both ORF1p and ORF2p are present in the nucleoli of some cells (25). A distinctive feature of ORF1p localization is its presence in large cytoplasmic foci similar in appearance to stress granules (SGs) or processing bodies (PBs).
Stress granules are discrete cytoplasmic aggregates which can be induced by a range of stress conditions, including heat shock, hypoxia, osmotic shock, oxidative stress, viral infection, and the overexpression of some cellular proteins (reviewed in references 1 and 14). Stalled preinitiation complexes are found in SGs and include the small, but not large, ribosomal subunits bound to translation eukaryotic initiation factors such as eIF2 and eIF3. Also present are poly(A)-binding protein (PABP), Ras-GAP SH3 binding protein (G3BP), and T-cell intracellular antigen 1 (TIA-1), as well as some other RNA-binding proteins that distinguish SGs from processing bodies (24, 40, 42, 78).
Processing bodies (PBs or GW bodies) are dynamic cytoplasmic compartments containing high concentrations of molecules involved in mRNA decay and translation inhibition. These include decapping proteins DCP1 and DCP2, exonuclease Xrn1, deadenylase CCR4, translation initiation factor eIF4E, and enzymes in the nonsense-mediated decay pathway (1). In mammalian cells, components of the RNA-induced silencing complex (RISC) have been found in PBs, including miRNAs, argonaute (Ago2), and GW182, an RNA-binding protein necessary for miRNA silencing (55, 69). PBs and SGs are related compartments, frequently overlap, and share and swap some components depending upon stress conditions in the cell. These observations have given rise to the concept of SGs as triage points where decisions are made as to whether dormant mRNPs will be transferred to processing bodies for degradation or returned to polysomes and the cytoplasmic pool of translating RNAs (42).
Here we demonstrate that ORF1p foci colocalize with markers of cytoplasmic SGs in both stressed and unstressed cells. Additionally, we show by mutation analysis that sequence modifications throughout ORF1p can profoundly change its localization, causing the protein to shift from the cytoplasm to the nucleus or nucleolus or altering the morphology or completely eliminating cytoplasmic foci. Based on interaction and colocalization studies, we also identified candidate non-L1 protein components of the L1 RNP. Most of these proteins are present with ORF1p in cytoplasmic foci. The discovery of L1 ORF1p together with polyadenylated RNA in stress granules suggests a mechanism whereby the cell may mitigate the potential mutagenic effects of retrotransposition by sequestering L1 RNPs and possibly targeting them for degradation.
MATERIALS AND METHODS
Cloning of plasmid constructs.
Open reading frames for DCP1b (NM_152640), eIF3B (NM_003757), glyceraldehyde-3-phosphate dehydrogenase (GAPDH; NM_002046), G3BP (NM_005754), HuR (NM_001419), PABPC1 (NM_002568), TIA-1 (NM_022173), and Xrn1 (NM_019001) were amplified by PCR from a human placental retroviral cDNA library (Clontech) and cloned into BamHI/SalI sites of pDsRed-Express-NI (Clontech). Coding sequences for DNA-binding protein A (DBP-A), DBP-B (YB-1), hnRNPA1, HSP70t, translocated-in-liposarcoma (TLS/FUS) protein, URH49, and 9G8 were amplified from pMyr two-hybrid cDNAs and cloned either with an N-terminus FLAG tag and three-residue linker (MDYKDDDDKAAG) into BamHI or NheI and PmeI sites of vector pcDNA6myc/hisB (Invitrogen) or as an N-terminal fusion to red fluorescent protein (RFP) in pDsRed-Express-NI. L1-RP ORF1 sequence (7) was cloned as either an N- or C-terminus fusion with green fluorescent protein (GFP) in SalI/BamHI sites of pEGFP-N1 or pC1-EGFP (Clontech), respectively, or with a C-terminal T7 tag and linker (AAGGMASMTGGQQMG) in pcDNA6myc/hisB, or a C-terminal tandem affinity purification (TAP) tag in the SalI/BamHI sites of pCMV-C-term-TAP. Tagged ORF1 sequences were also incorporated into full-length L1-RP and cloned in NotI/PmeI sites of the vector pcDNA6myc/hisB to generate constructs used in the immunoprecipitation experiments of Fig. 4, below. ORF1 truncation and point mutants were constructed by PCR techniques and cloned in pEGFP-N3. To generate pGEX 6P-ORF1-FL, ORF1 was cloned in BamHI/SalI sites of the pGEX 6P-1 glutathione S-transferase (GST) expression vector (Amersham Biosciences).
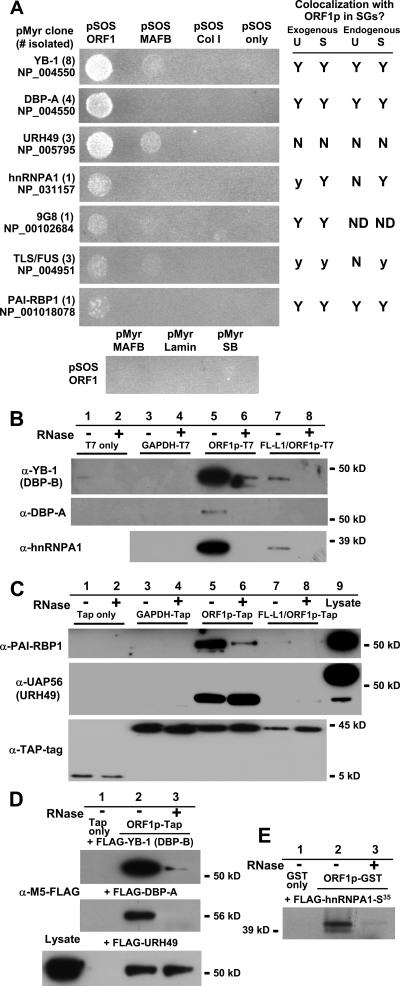
FIG. 4.
ORF1 protein associates with other cellular RNA-binding proteins. (A) Interaction of clones isolated from a human testes yeast Cytotrap two-hybrid library were confirmed by cotransfection in yeast with pSOS ORF1 or the following negative control plasmids: pSOS vector (containing the human Sos gene only), pSOS Col I (hSos gene and amino acids 148 to 357 of murine 72-kDa type IV collagenase), and pSOS MAFB (fusion of hSos gene and MAFB transcription factor). In some cases cotransfection with pSOS MAFB gave background growth. No interaction was detected when pSOS ORF1 was cotransfected with the following control plasmids: pMyr SB (myristylation signal fused with the Sos-binding protein gene), pMyr lamin C (myristylation signal fused with human lamin C, amino acids 67 to 230), and pMyr MAFB. Accession numbers for each isolated protein are shown. At right is a summary of data for colocalization with ORF1p in stress granules in unstressed and arsenite- or thapsigargin-stressed cells. Symbols: N, no significant overlap; Y, colocalization in a majority of cells; y, colocalization faint or obvious in only a small percentage of cells; ND, not determined. (B) T7-tagged ORF1p coimmunoprecipitates endogenous cold shock domain proteins YB-1 (DBP-B) and DBP-A and also hnRNPAI from 293T cytoplasmic extracts in an RNA-dependent manner. Although YB-1 has a predicted size of 36 kDa, it typically runs near 50 kDa, possibly due to posttranslational modifications (18). Construct FL-L1/ORF1p-T7 contains a full-length L1 with tagged ORF1 sequence. (C) TAP-tagged ORF1p coimmunoprecipitates endogenous PAI-RBP and a UAP56-related protein from 293T whole-cell lysates. Recovery of proteins from the affinity matrix was by TEV protease cleavage. The lower panel shows recovered TAP tag protein alone and TAP-tagged ORF1p recovered following TEV cleavage. (Prior to cleavage, ORF1p fused with full-length TAP-tag is 61 kDa.) FL-L1/ORF1-Tap is TAP-tagged ORF1 in full-length L1-RP. (D) Exogenously expressed FLAG-tagged YB-1 (DBP-B) and DBP-A coprecipitate with TAP-tagged ORF1p in an RNA-dependent manner. Coimmuniprecipitation of FLAG-tagged URH49 is not dependent upon RNA. (E) FLAG-tagged hnRNPA1 protein was labeled in reticulocyte lysate, incubated in vitro with Sepharose-bound GST-tagged ORF1p, and eluted with glutathione. Lysates were treated with 20 μg/ml RNase A or left untreated.
To construct the L1 reporter cassette construct 99-PUR-RPS-pBlaster1, the blasticidin S deaminase gene (bsd) from pcDNA6myc/hisA (Invitrogen) was excised with EspI-blunted XmnI and cloned in a modified pBS KS- vector lacking PvuII recognition sites. This construct was cut with SpeI and blunted, and into this site was inserted a minimal TK-poly(A) signal excised from the vector pOG-44-Cre (RsrII-blunted/SpeI-blunted). Intron 2 of the γ-globin gene was cloned in opposite orientation at the PvuII site within bsd, and this cassette was excised (EcoRV/SalI blunted) and inserted into a SmaI site introduced near the 3′ end of L1-RP within vector pBS KS- (JCC5-RPS). The L1-RP/bsd cassette was excised (NotI/ApaI blunted) and cloned into the vector 99-PUR (NotI/Sfi blunted). 99-PUR was constructed by combining an EcoRI/TthIII-blunted fragment from a modified version of vector pCEP4 (Invitrogen), lacking the cytomegalovirus (CMV) promoter, with an EcoRI-BamHI fragment from pPUR (Clontech). This hybrid construct contains the puromycin resistance gene.
GFP-GW182 (312-1709) was a gift from D. Bloch (Harvard Medical School), and pCMV-C-term-TAP was from E. Izaurralde (European Molecular Biology Laboratory). The Swimmer 1 (SW1) retrotransposon used to generate SW1 ORF1-EGFP was from D. Duvernell (Southern Illinois University).
Cell culture.
Human embryonic kidney HEK 293T cells and osteosarcoma 143Btk- cells (ATCC) were grown in high-glucose Dulbecco's modified Eagle's medium with 10% fetal bovine serum-glutamine-penicillin-streptomycin. Teratocarcinoma cell lines nTERA 2D1 and 2102Ep were gifts of J.V. Moran (University of Michigan) and P. K. Andrews (University of Sheffield, United Kingdom), respectively, and were grown in Dulbecco's modified Eagle's medium with minimal amino acids (Invitrogen). PERK/PKR/GCN2−/− mouse embryonic fibroblast (MEF) cells were a gift from D. Cavener (Penn State University). S/S and A/A MEFs were from R. Kaufman (University of Michigan). MEF wild-type and TIA-1−/− cell lines were a gift of N. Kedersha (Harvard University). All transfections used FuGENE6 (Roche).
Retrotransposition assay.
The retrotransposition cell culture assay was modified from reference 64. Wild-type L1 reporter construct 99-PUR-RPS-pBlaster1 (containing a blasticidin S deaminase gene reporter cassette) and mutant variants were transfected in 143Btk- cells in six-well plates. Antibiotic selection with puromycin (5 μg/ml) was begun 48 h posttransfection. On day 6 posttransfection, cells were expanded from six-well plates to T75 flasks. One week later, cells were trypsinized, counted, and reseeded in new T75 flasks in medium with blasticidin S (5 μg/ml). On day 22 posttransfection, cells were fixed and stained with Giemsa, colony numbers were scored, and the retrotransposition frequency was determined as the number of blasticidin-resistant colonies per number of transfected (puromycin-resistant) cells.
This lab and others have assayed full-length L1 reporter constructs containing ORF1 with C-terminal tags and have found them to retain retrotransposition competence in cell culture. Tagging ORF1 at the N terminus abolishes retrotransposition (46; unpublished data).
Antibodies.
Purified AH40.1 polyclonal antibody against ORF1 protein was a gift from T. Fanning (Armed Forces Institute of Pathology, MD) and M. Singer (Carnegie Institution of Washington). Affinity tag antibodies included anti-T7 tag (Novagen), anti-TAP tag (Open Biosystems), and anti-FLAG tag (Sigma). Other primary antibodies and their sources were anti-Ago2 (Z. Mourelatos, University of Pennsylvania), anti-DBP-A and anti-DBP-B (YB-1; K. Kohno, University of Occupational and Environmental Health, Japan), anti-eIF3η (N-20; Santa Cruz Biotechnology, Inc.), anti-FMRP (clone 1C3; Chemicon), anti-GW182 (M. Fritzler, University of Calgary), anti-hnRNPA1 (clone 4B10; G. Dreyfuss, University of Pennsylvania), anti-LAMP2 (H4B4; Developmental Studies Hybridoma Bank, University of Iowa), anti-PABPC1 (clone 10E10; G. Dreyfuss), anti-plasminogen activator inhibitor RNA-binding protein 1 (PAI-RBP1; J. Peluso, University of Connecticut), anti-PDI (Stressgen Bioreagents), anti-TIA-1 (C-20; Santa Cruz), anti-TLS (BD Transduction Laboratories), and anti-UAP56 (M. Green, University of Massachusetts Medical Center). Donkey Cy2- or Cy3-conjugated and horseradish peroxidase-conjugated anti-mouse and anti-rabbit secondary antibodies were from Jackson ImmunoResearch Labs. Horseradish peroxidase-conjugated anti-chicken immunoglobulin Y (IgY) antibody was from GenWay Biotech. Lysotracker Red DDN-99 is a product of Molecular Probes, Invitrogen.
Immunofluorescence.
Cells grown on poly-l-lysine-treated coverslips were washed twice with Dulbecco phosphate-buffered saline (DPBS) for 5 min and fixed with cold 3% DPBS-paraformaldehyde (pH 7.3) for 20 min at 4°C, followed by washing and permeabilization for 10 min at 4°C in DPBS-0.2% Triton-X. Cells were incubated with protein blocking agent (Immunotech) for 15 min and overnight at 4°C with primary antibodies in DPBS-5% donkey serum-0.1% Triton-X. The next day, cells were washed three times with DPBS and incubated for 2 h with donkey Cy2- or Cy3-conjugated secondary antibodies. Coverslips were mounted with mounting medium (KPL Laboratories).
For polyadenylated mRNA detection, following paraformaldehyde fixation, cells on coverslips were incubated overnight in 70% ethanol at 4°C, dehydrated through a 80%, 90%, 100% ethanol series, and dried on a heat block. Coverslips were incubated overnight at 37°C with 100 ng of a 45-nt Cy3-conjugated poly(dT) probe or Cy3-labeled antisense DNA probe recognizing a region of the L1 3′ UTR (Integrated DNA Technologies). Hybridization buffer contained 10% dextran sulfate, 2 mM vanadyl-ribonucleoside complexes, 40 μg Escherichia coli tRNA, 25 μg salmon sperm DNA, 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 15% formamide, and 1 μl RNasin (Promega). Cells were washed twice for 30 min in 2× SSC-15% formamide at 37°C, followed by primary antibody (1 h) and secondary antibody (2 h) incubation as required.
Slides were examined with either a Leica DR RE microscope with a TCS SP confocal imaging spectrophotomer using Leica TCSNT version 1.6.587 software or a Nikon E600 microscope equipped with bright-field, Nomarski, and fluorescence optics (Fast 1934 CCD camera; QImaging, QICAM, Canada) and IP Lab imaging software (Scanalytics Corp., BD Biosciences).
Immunoprecipitation.
Cells were harvested in DPBS-2 mM EDTA, and the cell membrane was lysed on ice for 20 min (20 mM HEPES, 20% glycerol, 200 mM KCl, 0.5 mM EDTA, 0.1% NP-40, with protease inhibitor cocktail [Sigma], 1 mM phenylmethylsulfonyl fluoride, and 0.5 mM dithiothreitol). Total cell lysates were obtained by sonication. Lysate was cleared by centrifugation at 11,000 rpm. Extracts were treated in the presence or absence of 20 μg/ml RNase A (Roche). All TAP tag immunoprecipitations were single round. Following overnight incubation at 4°C with cross-linked IgG beads (Sigma), elution was by cleavage with tobacco etch virus protease (Invitrogen) in cleavage buffer (10 mM Tris-Cl, pH 8, 150 mM KCl, 0.1% NP-40, 0.5 mM EDTA, 1 mM dithiothreitol).
For the GST pull-down assay (see Fig. 4E, below), construct pGEX 6P-ORF1-FL was induced in BL21-CodonPlus(DE3)-RIPL (Stratagene) bacteria, the pellet was lysed, and recombinant ORF1p protein was purified on glutahione-Sepharose 4B resin according to the manufacturer's protocols (Amersham Biosciences). Nine μl of FLAG-tagged hnRNPA1 protein labeled with 35S (TNT T7 coupled reticulocyte lysate system; Promega) was incubated for 3 h with bound GST-tagged ORF1p, washed five times with PBS, and eluted with 20 mM glutathione.
Two-hybrid analysis.
A Stratagene Cytotrap system human testis library was screened according to the manufacturer's instructions with the full-length ORF1 sequence in pSOS vector. Approximately 1.2 × 1010 plasmids were used to transform the CDC25α yeast strain. Putative ORF1p-interacting library clones were tested for noninteraction with control expression constructs pSOS alone, pSOS MAFB, and pSOS ColI.
RESULTS
L1 ORF1 protein is predominantly cytoplasmic and concentrates in discrete foci.
Previously we reported that both ORF1 and ORF2 proteins were predominantly cytoplasmic when overexpressed in the MVA/T7RP system but also targeted nucleoli in some cells (25). Here we detected ORF1p, tagged on either its N or C terminus with GFP and expressed either alone or together with ORF2p, in nucleoli in about 5% of cells (Fig. 1A; see also Fig S1A in the supplemental material). Faint general nuclear staining was also seen, but only a small number of cells showed nuclear concentration.
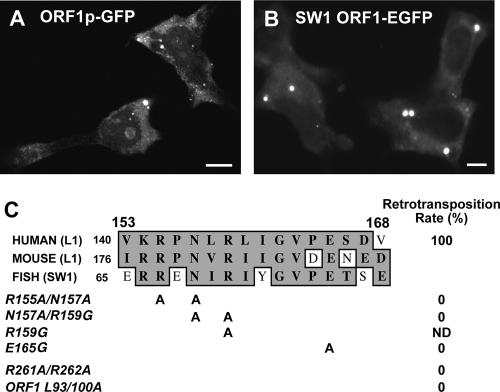
FIG. 1.
(A) ORF1p tagged on the C terminus with enhanced GFP is predominantly cytoplasmic but is also present in nuclei and nucleoli of some cells. (B) GFP-tagged ORF1p from the fish retrotransposon SW1 is cytoplasmic with punctate foci in human 143Btk- cells. Nucleolar localization of this protein was not observed. (C) Phylogenetic alignment of internal sequence from ORF1p and the location of point mutations. Mutations were also introduced into a retrotransposition-competent L1 (L1-RP) and tested in a cell culture retrotransposition assay. Retrotransposition rates were compared with that of wild-type L1-RP (right).
A dominant feature of ORF1p was its concentration in cytoplasmic foci that fluoresced intensely against a background of diffuse cytoplasmic expression. These phase-dense particles varied in size and number, although there were usually <10 per cell. This pattern is well-conserved through evolution, since GFP-tagged ORF1 protein from SW1, a related non-long terminal repeat (non-LTR) retrotransposon found in some teleost fish species (15), was similarly distributed (Fig. 1B). Martin and Branciforte (59) also reported punctate cytoplasmic staining of endogenous mouse L1 ORF1p in F9 cells and proposed that the speckles were RNP aggregates. Human ORF1p with a C-terminal T7 epitope tag showed the same pattern of distribution as ORF1p-GFP when assayed by immunofluorescence.
A central region of ORF1p is important for cytoplasmic distribution.
To identify domains responsible for subcellular localization, we generated a series of truncation and point mutants of ORF1p, fused these upstream of GFP, and assayed for expression following plasmid transfection in human osteosarcoma 143Btk- and embryonic kidney 293T cells. Our data (summarized in Table 1; see also Fig. S1 in the supplemental material) indicate that the N-terminal-third of the protein contributes to cytoplasmic retention of ORF1p, since residues 1 to 130 were largely cytoplasmic but did not form foci. Fragment 1-168 also failed to form foci but was cytoplasmic in some cells and nuclear in others, suggesting that sequence downstream of the zinc finger domain (residues 90 to 130) can direct ORF1p to the nucleus but not necessarily the nucleolus. Hence, nuclear and nucleolar localization can be uncoupled. Removing portions of both the N and C termini unmasked this activity and caused the truncated fragments to enter the nucleus.
TABLE 1.
Subcellular localization patterns of ORF1p-GFP truncation and point mutants expressed in untreated 143Btk- and 293T cells
| Mutant | Localizationa
|
|||
|---|---|---|---|---|
| Cytoplasmic | Cytoplasmic foci | Nuclear | Nucleolar | |
| 1-339 (wild type) | *** | *** | * | * |
| 1-130 | *** | X | * | X |
| 1-168 | ***/* | X | */*** | ND |
| 1-270 | *** | *(1) | * | * |
| 51-339 | *** | *** | * | X |
| 89-339 | *** | ** | * | X |
| 51-168 | * | X | *** | Ex |
| 89-270 | * | * | *** | Ex |
| 131-339 | * | * | *** | Ex. |
| 169-270 | * | X | *** | Ex/* |
| 195-270 | * | X | *** | Ex/* |
| 225-339 | ** | X | *** | Ex/* |
| R155A/N157A | *** | *** | * | * |
| N157A/R159G | *** | X | ** | * |
| R159G | *** | *(1) | * | X |
| E165G | *** | *** | * | *** |
| L93/100/114A | *** | **(1) | * | * |
| R261/262A | *** | **(1) | * | X |
Symbols: X, not observed; Ex, excluded; (1), foci altered in morphology from wild type; ***, predominant; **, less frequent; *, infrequent; ND, not determined.
Our data also indicate that a domain that overlaps residue 168 affects foci formation. While fragments 51-168 and 169-270 were exclusively nuclear without foci, longer constructs 89-270 and 131-339 were nuclear but also retained foci remnants in some cells. We next tested the effects of point mutations introduced between residues 154 to 167, a sequence highly conserved between human L1, mouse L1, and fish SW1 retrotransposons (Fig. 1C). Foci formation was affected by changes within this central domain. While the double point mutation R155A/N157A had no obvious effect on ORF1p localization, N157A/R159G completely abolished foci, while the single mutation R159G severely reduced their number and altered their morphology, being paler and larger in size. Interestingly, point mutation E165G did not inhibit foci formation and cytoplasmic staining but increased the percentage of 143B and 293T cells having visible nucleolar staining by four- to fivefold. Introducing these residue changes in a full-length retrotransposition-competent L1 containing a blasticidin resistance gene reporter cassette eliminated its retrotransposition in a cell culture assay (64) (Fig. 1C).
We also considered that the leucine zipper domain might be important for foci formation. Changing three leucines to alanines (L93/100/114A) did not affect cytoplasmic localization, but foci were observed in fewer cells and often differed in morphology. We observed a similar foci pattern for R261A/R262A, a double point mutation known to abolish retrotransposition in cell culture (64). Mutating leucines of the leucine zipper also inactivated L1 in cell culture.
We failed to identify cis-acting cytoplasmic, nuclear, or nucleolar localization signal sequences in ORF1p. No nuclear localization signal was predicted in ORF1 by either the PredictNLS or WoLF PSORT server (12, 32). Moreover, despite the presence of a strong nuclear export signal predicted by the program NetNES (residues 213 to 221 [47]), export of ORF1p is independent of the chromosomal region maintenance 1 (CRM1) pathway, as determined by its lack of sensitivity to the CRM1 inhibitor leptomycin B (25; unpublished data). Although such signals may exist, we suggest that protein secondary structure, likely mediating protein or RNA binding, is critical for wild-type distribution of ORF1p. This protein structure may be easily disrupted by mutation throughout the length of the protein, altering macromolecule binding and normal targeting of the ORF1p RNP. For example, data suggest that RNA binding is required for nucleolar targeting. Treating 143B or 193T cells with the transcription inhibitor actinomycin D abolished visible nucleolar localization of both GFP-tagged wild-type ORF1p and the E165G mutant. Cytoplasmic patterning and foci formation remained unchanged (not shown).
ORF1p cytoplasmic foci localize with markers of stress granules.
Martin and Branciforte (59) reported that punctate cytoplasmic staining of endogenous mouse ORF1p was not associated with the Golgi apparatus, lysosomes, or endoplasmic reticulum (ER). Similarly, cytoplasmic foci generated by tagged human ORF1p did not align with endogenous markers for ER or lysosomes (see Fig. S2 in the supplemental material).
ORF1p was cotransfected with fluorophore-tagged marker proteins and examined for colocalization in 143Btk- and 293T cells. As expected, GFP-tagged ORF1p did not colocalize in cytoplasmic foci with coexpressed red fluorescent protein alone (not shown), RFP-tagged GAPDH, or RFP-tagged heat shock protein 70 homolog HSP70t (Fig. 2A and B).
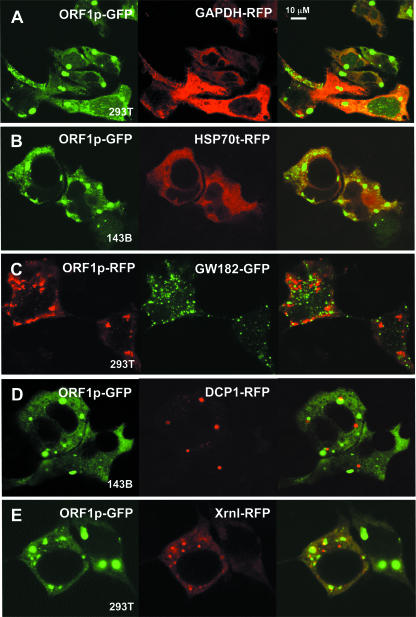
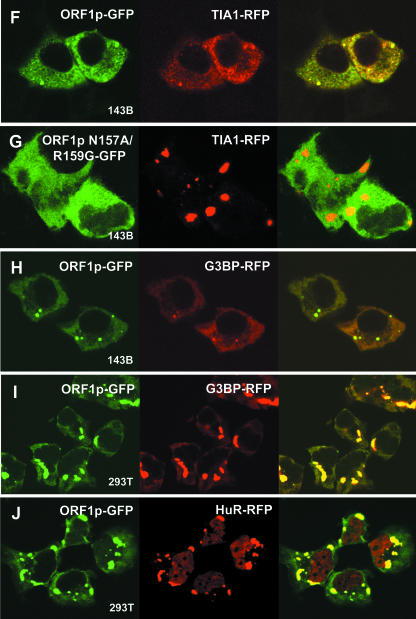
FIG. 2.
ORF1p colocalizes with marker proteins of stress granules. Fluorescent-tagged ORF1p was coexpressed in 143Btk- or 293T cells with fluorescence-tagged control proteins (A and B), marker proteins for PBs (C to E), and marker proteins for SGs (F to J). (G) Cells were cotransfected with TIA1-RFP and ORF1p-GFP containing a double point mutation, N157A/R159G, that prevents formation of cytoplasmic foci. Cells were imaged by confocal microscopy at ×100 magnification. Bar, 10 μm.
We next considered that ORF1p foci might be cytoplasmic RNA granules, either processing bodies or stress granules (Table 2). To test for PB localization, we examined fluorophore-tagged ORF1p coexpressed with GFP-tagged GW182 (Fig. 2C) or RFP-tagged decapping protein DCP1 (Fig. 2D), both bona fide markers of PBs (1). Foci formed by these PB marker proteins tended to be smaller and did not generally overlap, although they were frequently juxtaposed with ORF1p foci. PBs and SGs are often found in close proximity (42). Close association of foci formed by RFP-tagged Xrn1 and ORF1p-GFP was more evident than that seen for DCP1 and GW182 (Fig. 2E). Kedersha et al. (42) have reported that Xrn1 is found in PBs in unstressed cells but partially or completely relocalizes to SGs in stressed cells.
TABLE 2.
Exogenous or endogenous PB and SG marker proteins and their colocalization with ORF1p in unstressed and stressed cells
| Marker group and name | Protein function | Exogenousa
|
Endogenousa
|
||
|---|---|---|---|---|---|
| Unstressed | Stressed | Unstressed | Stressed | ||
| PB marker proteins | |||||
| GW182 | miRNA-mediated mRNA decay | N | N | N | N |
| DCP1/2 | mRNA decapping and decay | N | N | ND | ND |
| Xrn1 | 5′-3′ exonuclease, mRNA decay | N/y | N/y | ND | ND |
| SG marker proteins | |||||
| TIA-1 | Splicing modulation, translation repression | Y | Y | y | Y |
| G3BP | Signal transduction, RNA metabolism | Y | Y | ND | ND |
| eiF3 complex | 40S binding, translation initiation | Y | Y | y | Y |
| HuR | RNA transport and stability | Y | Y | ND | ND |
| PABPC1 | Translation initiation, mRNA decay | ND | ND | N | Y |
Symbols: N, no significant overlap; Y, colocalization in a majority of cells; y, colocalization faint or present in a small number of cells; ND, not determined.
We next tagged protein markers of SGs and coexpressed these with ORF1p-GFP. RFP-tagged TIA-1, G3BP, and HuR (ELAV1), an AU-rich element-binding protein (21), all colocalized with ORF1-EGFP protein in many 143B cells under normal conditions (Fig. 2F and H; Table 2). Colocalization was even more dramatic in 293T cells, in which protein expression was higher and foci were larger (Fig. 2I and J). No colocalization to foci was seen when the point mutant N157A/R159G-GFP was expressed with these stress granule marker proteins (Fig. 2G).
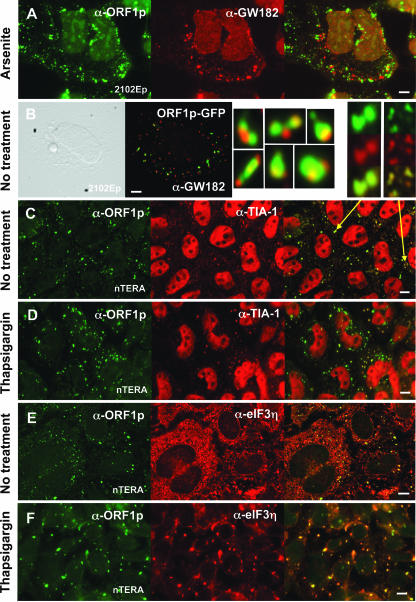
It has been reported that simply overexpressing the stress granule-associated proteins G3BP or TIA-1 causes their self-aggregation and triggers SG formation (24, 78). We considered that this might also be the case for endogenous L1 ORF1p and determined its localization in germ cell-derived cell lines that express L1 RNA and ORF1p at high levels (29, 48). The well-characterized AH40.1 polyclonal antibody detects endogenous ORF1 protein strongly as a single dominant band in cytoplasmic extracts from the human embryonal germ cell teratocarcinoma-derived cell lines nTERA 2D1 and 2102Ep but very weakly in extracts from non-germ cell lines when assayed by Western blotting (48) (see Fig. S3A in the supplemental material). We now report that in 2103Ep and nTERA cells this antibody detects a general cytoplasmic localization of endogenous ORF1 protein with concentrations in cytoplasmic aggregates similar to those seen with transfected constructs (Fig. 3; see also Fig. S3B in the supplemental material).
FIG. 3.
ORF1p is targeted to stress granules in both unstressed and stressed teratocarcinoma cells. The distribution of tagged or endogenous ORF1p in teratocarcinoma cells was compared with that of endogenous marker proteins of PBs (A and B) and SGs (C to H) and a Cy3-labeled poly(dT) probe that detects polyadenylated RNA (I to K). Enlarged images of individual cytoplasmic foci are shown beside panel B and above panel C. The poly(dT)-Cy3 probe clearly detects polyadenylated RNA in stressed (J and K) but not unstressed (I) cells. Some cell groups were chemically stressed by treatment with sodium (meta)arsenite (0.75 mM; 50 min) or thapsigargin (1.5 μM; 2 h) immediately prior to fixation. Bar, 10 μm.
We examined the expression of endogenous marker proteins and ORF1p under both unstressed and stressed cell conditions. To examine association with PBs, endogenous ORF1p was costained with an antibody against GW182 (Fig. 3A). As previously reported, arsenite-induced oxidative stress significantly increased the number of cytoplasmic PBs (42). We did not find general overlap of ORF1p and GW182 protein under either stressed or unstressed conditions, although frequent juxtaposition of ORF1 aggregates and PBs was evident (Fig. 3B and enlarged images).
TIA-1 is a predominantly nuclear protein that rapidly localizes to cytoplasmic stress granules when cells are treated with arsenite or with thapsigargin, a compound that blocks calcium uptake into the ER lumen and triggers the unfolded protein response. Colocalization of endogenous TIA-1 with both GFP-tagged and endogenous ORF1p in many unstressed cells was obvious but faint (Fig. 3C and enlarged images). Upon chemical treatment of teratocarcinoma cells, a dramatic realignment of TIA-1 with ORF1p in large cytoplasmic granules occurred (Fig. 3D). However, TIA-1 expression is not required for foci formation, as no obvious differences in localization patterns were seen for GFP-tagged ORF1 transfected in mouse embryonic fibroblast cell lines derived from wild-type and TIA-1 knockout mice (51).
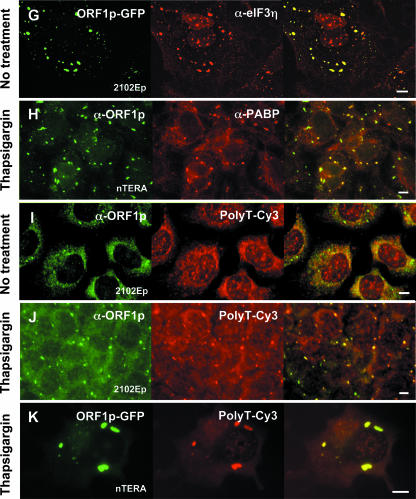
As with TIA-1, colocalization of ORF1p and endogenous elongation initiation factor 3 (eIF3) (Fig. 3E to G) was seen in both unstressed and, to a greater degree, in stressed cells. On the other hand, colocalization of ORF1p with PABP, another bona fide marker of SGs, was not obvious in unstressed cells (not shown) but increased dramatically under stress conditions (Fig. 3H). Similarly, a Cy3-conjugated 45-nt poly(T) DNA probe clearly detected polyadenylated RNA in ORF1p (Fig. 3I to K) or TIA-1 (not shown) foci only under induced stress conditions. Probe signal disappeared when cells were pretreated with RNase A (not shown). This suggests that the amount of polyadenylated mRNA present in ORF1p foci is low under normal cellular conditions but that significant populations of RNPs are shunted to these sites following stress. We failed to specifically detect L1 transcripts using a Cy3-conjugated DNA probe recognizing sequence within the L1 3′ UTR. The tendency of ORF1p to bind its own encoding transcript (cis preference) and its predicted ability to coat L1 RNA (reviewed in reference 2) likely mean that L1 RNA is far less abundant than ORF1p in stress granules and more difficult to detect.
Inhibition of translation is not a prerequisite for ORF1p aggregate formation.
Stress granule formation occurs following initiation of cell stress and is usually preceded by phosphorylation of translation initiation factor eIF2α. Phosphorylation at Ser51 increases the binding affinity of eIF2α for eIF2B, a guanine nucleotide exchange factor that charges the eIF2-GTP-tRNAMet ternary complex. This prevents eIF2B recycling and blocks polysome assembly and further rounds of translation. We assayed whether phosphorylation of the translation initiation factor eIF2α is necessary for ORF1p foci formation. GFP-tagged ORF1p was expressed in MEF TKO cells carrying deletions of three of the four known eIF2α kinases: the ER-localized kinase PERK, the double-stranded RNA-activated protein kinase PKR, and GCN2, the mammalian homolog of yeast Gcn2p (36). (A fourth kinase, heme-regulated inhibitor kinase HRI, is most abundant in erythroid cells.) No obvious difference in ORF1p distribution was seen between MEF TKO cells and parental MEF +/+ cells (see Fig. S4A and B in the supplemental material). On the other hand, MEF TKO cells would not form stress granules when treated with arsenite or thapsigargin (see Fig. S4E and F in the supplemental material). Similarly, while A/A MEF cells containing a mutant nonphosphorylatable form of eIF2α could not generate SGs when exposed to chemical stress (63) (data not shown), ORF1p-GFP localization in this mutant line was not obviously different from that seen in wild-type MEF S/S cells (see Fig. S4C and D in the supplemental material). Thus, ORF1p forms SGs independent of eIF2α phosphorylation. Other authors have also reported a similar phenomenon. As noted above, some proteins, such as TIA1 and G3BP, act downstream of eIF2 phosphorylation and cause stress granule formation when expressed at high levels (24, 78). Furthermore, pateamine A, a translational inhibitor that targets eIF4A activity, has been shown to induce SGs independently of eIF2α (13).
Cells treated with arsenite and other inducers of stress fail to assemble SGs in the presence of emetine or cycloheximide, drugs which stabilize polysomes and prevent translation (41). However, ORF1p remained unaffected under the same conditions, further evidence that general inhibition of cellular translation is not a precondition for their formation (see Fig. S4G and H in the supplemental material).
RNA-binding proteins colocalize with ORF1p in stress granules.
To identify proteins associated with L1 retrotransposons, we employed the yeast two-hybrid CytoTrap system (Stratagene) to screen a human testes plasmid cDNA library using full-length ORF1p as bait. This system has the advantage over conventional GAL4 and LexA two-hybrid systems of assaying for interactions in the cytoplasm, the main site of ORF1p localization. Most proteins identified were RNA-binding proteins and were confirmed for interaction in the Cytotrap system by individual screening against ORF1p bait and several control proteins. Seven interacting proteins were selected for further analysis by ORF1p coimmunoprecipitation and colocalization (Fig. 4A).
Clones most frequently isolated from the two-hybrid screen were DBP-B (YB-1) and DBP-A, two “cold shock domain” nucleic acid-binding proteins involved in a wide range of cellular functions, including DNA repair, regulation of transcription and translation, and stress response. Both are predominantly cytoplasmic, and while DBP-B is ubiquitously expressed in many somatic and cancer tissues, DBP-A expression is generally limited to heart and skeletal muscle, decidual cells, spermatocytes, and some carcinomas (44). Multiple clones were also isolated for URH49 (also called DDX39), a DExD/H-box protein that is 90% identical to the nuclear RNA helicase UAP56, which plays important roles in both mRNA splicing and nuclear export. It is likely that these two proteins share similar roles in the cell, although URH49 is expressed at significantly higher levels in testes (68). The CytoTrap system also identified as potential interactors with ORF1p several clones for TLS/FUS protein and single clones for hnRNPA1, the SR (arginine/serine-rich) shuttling protein 9G8, and plasminogen activator inhibitor RNA-binding protein 1 (PAI-RBP1). hnRNPA1 is an abundant RNP protein that associates with pre-mRNA, nuclear-spliced mRNA, and cytoplasmic mRNA. 9G8 functions in the export of both spliced and intronless RNA and operates in the same NXF1/TAP-dependent export pathway as UAP56 (33). TLS/FUS plays roles in both spliceosome assembly and transcription. PAI-RBP1 was originally identified as binding PAI1 mRNA and playing a role in its stability (28). Subsequently, its Drosophila melanogaster homolog, VIG (encoded within an intron of the Vasa gene), was identified as part of RISC in that species (9).
Antibodies directed against either a TAP tag or a T7 tag fused to the C terminus of ORF1p coimmuniprecipitated endogenous DBP-A, YB-1, hnRNPA1, and PAI-RBP1 from 293T cells (Fig. 4B and C). These associations diminished when cell lysates were pretreated with RNase A, suggesting that RNA forms a scaffold upon which these other proteins are assembled in RNPs with ORF1p. When tagged ORF1p was expressed from a full-length L1, coprecipitated proteins were still detected, although in significantly lower amounts. Diminished levels of ORF1p were expressed from a bicistronic RNA (Fig. 4C, lower panel), and this may be due in part to cellular toxicity of overexpressed ORF2p (22, 25). Similarly, when T7-tagged ORF1p was coexpressed with FLAG-tagged DBP-A or YB-1 and analyzed with anti-FLAG antibody, coimmunoprecipitated proteins were isolated, and this interaction was abolished with RNase A treatment (Fig. 4D). GST-tagged ORF1p was also able to precipitate hnRNPA1 protein that had been translated and labeled with 35S in reticulocyte lysate (Fig. 4E).
TAP-tagged ORF1p selectively coimmunoprecipitated an endogenous protein product slightly smaller than expected for URH49 and UAP56 proteins when assayed with the UAP56 polyclonal antibody generated by Fleckner et al. (19) (Fig. 4C). This minor band is also visible in the Western blot analyses presented by those authors. The nature of this smaller product is unknown, although splicing variants and posttranslational modifications of URH49 have been reported (76). Both the efficiency of ORF1p-TAP purification and the amount of coimmunoprecipitate increased slightly in the presence of RNase A, suggesting both that removing bound RNA may alter the conformation of ORF1p and that ORF1p interacts directly with a UAP56-related protein. Similarly, exogenous FLAG-tagged URH49 was coimmunoprecipitated by TAP-tagged ORF1p, and this association was not altered by incubation with RNase A (Fig. 4D).
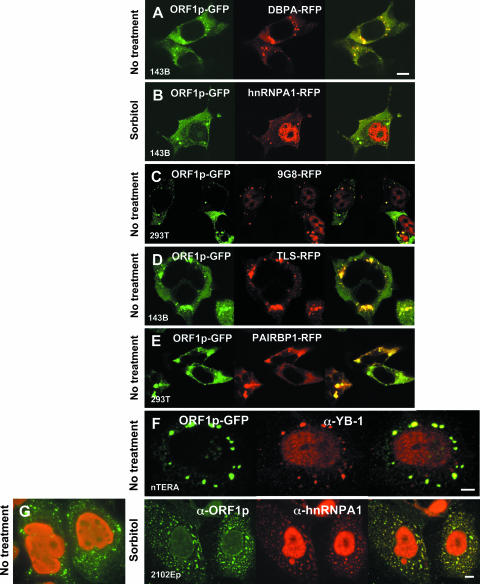
Next, proteins detected by the two-hybrid screen were tagged on their C termini with RFP and coexpressed with ORF1p-GFP to assay for coalignment (results are summarized in Fig. 4A). DBP-A-RFP colocalized with ORF1-GFP in foci that tended to be larger and more diffuse than those formed by ORF1p-GFP alone (Fig. 5A). Similarly, ORF1-GFP colocalized with endogenous DBP-A (not shown) and YB-1 (Fig. 5F) in large foci in a significant proportion of teratocarcinoma cells. This alignment was apparent in both stressed and unstressed cells. Gallois-Montbrun (20) recently reported that YB-1, in association with the antiretroviral factor APOBEC3G, relocalizes from PBs to SGs when cells are subjected to heat shock stress.
FIG. 5.
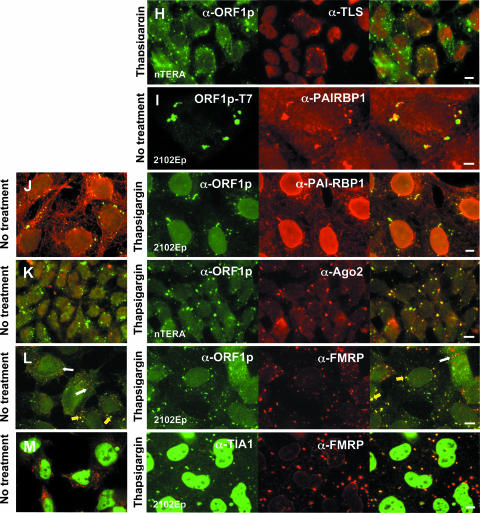
ORF1p colocalizes with other RNA-binding proteins in stress granules. (A to E) GFP-tagged ORF1p was coexpressed in 143Btk- or 293T cells with RFP-tagged proteins identified from the two-hybrid screen. Cells were treated with 600 mM sorbitol or left untreated. (F to L) Localization of endogenous, GFP-fused, or T7 epitope-tagged ORF1p with endogenous proteins in teratocarcinoma cells. Cells were left untreated or treated with thapsigargin or sorbitol 2 h prior to fixation. In panel L, yellow arrows show alignment of FMRP with ORF1 protein in SGs. White arrows show smaller foci formed by FMRP, presumably PBs, which do not align with ORF1p. (M) Coexpression of endogenous FMRP and TIA-1 proteins in treated and untreated teratocarcinoma cells. Cells were imaged with a Nikon E600 microscope at ×60 magnification or confocal microscopy at ×100 magnification. Bar, 10 μm.
Guil et al. (26) reported that hnRNPA1 relocalizes from the nucleus to cytoplasmic stress granules, but not processing bodies, in 100% of HeLa or murine 3T3 cells subjected to osmotic (sorbitol-induced) stress and 40% of cells subjected to heat shock or oxidative (arsenite-induced) stress. Sorbitol treatment also induced close alignment of ORF1p and hnRNPA1 when either flourophore-tagged or endogenous proteins were assayed (Fig. 5B and G). On the other hand, URH49 is a predominantly nuclear protein and when tagged with RFP, or assayed endogenously, it was not generally found in the cytoplasm with ORF1p, even when cells were stressed (not shown). Similar to the cold shock domain proteins, RFP-tagged 9G8, a nucleo-cytoplasmic shuttling protein, was frequently found in foci formed by ORF1p-GFP in 293T and 143B cells, whether cells were chemically stressed or not (Fig. 5C). On the other hand, although transfected RFP-tagged TLS/FUS, another shuttling protein, was present with ORF1p-GFP in foci of some unstressed 143B and 293T cells (Fig. 5D), endogenous TLS/FUS was almost exclusively nuclear in unstressed nTERA cells and not observed in foci formed by ORF1p (not shown). When nTERA cells were treated with thapsigargin, a small number of cells (less than 0.5%) showed distinctive aggregation of TLS/FUS with ORF1p proteins (Fig. 5H). The significance of this rare alignment is unclear.
PAI-RBP1 was detected in the nucleus, perinuclear region, nucleoli, and cytoplasm, as reported by Lemos and Kobarg (49). Those authors have also identified nuclear colocalization with p80-coilin-positive coiled bodies. In both stressed and untreated cells, tagged and endogenous PAI-RBP1 aggregated in densely staining cytoplasmic foci that colocalized with both plasmid-expressed and endogenous ORF1p (Fig. 5E, I, and J).
We next examined expression of endogenous argonaute 2 (Ago2) protein in 2102Ep and nTERA 2D1 cells using a well-characterized antibody (65). Ago2 is a component of RISC and mediates RNA cleavage targeted by miRNAs and siRNAs. Ago proteins and miRNAs localize to processing bodies (54, 55, 67, 70), but under stress conditions Argonaute proteins also undergo miRNA-dependent localization to stress granules (50). We found that Ago2 colocalized with ORF1p in thapsigargin-treated cells but not obviously in untreated cells (Fig. 5K). Under untreated conditions, Ago2 was generally distributed throughout the cytoplasm, and discrete foci consistent with P-bodies were not evident in many teratocarcinoma cells. This is not surprising, since it has recently been reported that only 1.3% of Ago2 resides in P-bodies and that these may disappear under some serum conditions (50, 80).
We also tested colocalization of ORF1p and the human fragile X protein FMRP, which has been reported to translocate from polyribosomes to SGs following arsenite treatment (43, 62a). Both VIG and the Drosophila FMRP ortholog, dFMR, associate with RISC and miRNAs in S2 cells (9). In mammalian cells, FMRP has been reported to be part of a complex containing Ago2 protein and miRNAs (37). Like PAI-RBP1, FMRP colocalized with ORF1p in both unstressed and stressed cells, although the number of cells in which this association was apparent increased significantly upon chemical treatment. Interestingly, under both conditions a minor fraction of cells displayed numerous small foci that did not align with ORF1p granules (Fig. 5L). FMRP also aligns with TIA-1 foci under stress conditions (Fig. 5M).
In summary, L1 ORF1p forms cytoplasmic aggregates whether cells are artificially stressed or not. Most of the proteins identified in the two-hybrid screen also associate with ORF1p in stress granules in chemically stressed and in some cases unstressed cells. Some proteins present with ORF1p in foci are integral components of the RNA interference RISC pathway.
DISCUSSION
Both ectopically expressed and endogenous ORF1p intensely accumulate in cytoplasmic foci. ORF1p is also faintly present in the nucleus, and a small percentage of cells show nucleolar targeting. In double-labeling experiments, the cytoplasmic foci costain with protein markers of stress granules in both stressed and unstressed conditions, but chemical induction of stress significantly increases the intensity of costaining. Furthermore, due to the presence of both PABP and polyadenylated RNA (Fig. 3H to K), we infer that ORF1p cosegregates with a large pool of mRNAs in SGs under stress conditions. Although we have yet to detect L1 transcripts, the presence of ORF1p in SGs and its cis preference for binding its own encoding RNA (17, 82) suggest that L1 RNA is also in stress granules.
Subcellular distribution of proteins is determined either by specific signal sequences or by interactions with other macromolecules. We have failed to isolate short specific cis-acting sequences within the ORF1 protein that control nucleocytoplasmic localization or stress granule formation. Rather, changes throughout the protein, and especially in its central conserved region, profoundly affect localization. This is dramatically demonstrated by the fact that a point mutation, R159G, abolishes foci formation, while E165G greatly increases nucleolar targeting. These data suggest that protein secondary structure and the ability to bind other macromolecules may be key determinants of ORF1p distribution in the cell.
Both L1 ORF1 and ORF2 proteins localize to nucleoli. What role might the nucleolus play in L1 retrotransposition? In addition to ribosome biogenesis, the nucleolus is associated with RNP assembly and RNA export for some viruses and LTR retrotransposons. Perhaps this is also the case for L1 RNPs. In yeast, the integrase protein of Ty3 and the Gag protein of Tf1 retrotransposons target the nucleolus (52, 77). One class of non-LTR elements, the site-specific R1 and R2 retrotransposons of arthropods, have evolved to insert into 28S rRNA genes at nucleoli (83).
We previously suggested (25) that transit of L1 RNPs through nucleoli might explain the unexpected discovery by Buzdin et al. (8) of numerous chimeras between L1s and small RNA gene sequences in the human genome, including L1 fused with U3, U5, U6, and 5S sequences, as well as 5S-Alu, 7SL-Alu, Alu-mRNA, and U6-mRNA hybrids. All derive from small RNAs known to traffic the nucleolus. A de novo insertion of an L1/U6 chimera was isolated from the retrotransposition cell culture assay and was likely generated by RNA template switching of ORF2p during reverse transcription (23). Recently, hundreds of small nucleolar RNA-derived retrogenes having the hallmarks of L1-mediated insertion were identified in mammalian genomes (57, 81).
Using two-hybrid screens we isolated seven RNA-binding proteins that associate with ORF1p. Of those tested in coimmunoprecipitation assays, only interaction with the DEAD-box protein URH49 was retained following treatment with RNase A, showing that the other proteins were detected because they simultaneously bind the same RNA transcript with ORF1p. All except URH49 were found with ORF1p in large cytoplasmic foci. Two of the ORF1p-interacting proteins, hnRNPA1 and YB-1, have recently been identified in stress granules (20, 26). The associations of DBP-A, PAI-RBP1, 9G8, and TLS/FUS with these granules are reported here for the first time. We speculate that a number of stress granule proteins, including but not limited to those discovered here, play a role in the L1 life cycle. These proteins may accompany L1 RNPs to stress granules.
Many of the proteins isolated from the two-hybrid screens mutually interact and have roles in mRNA transport. For example, hnRNPA1 is an abundant nuclear protein involved in splicing and export of several mammalian and viral pre-mRNAs. TLS/FUS associates in vivo with hnRNPA1 and YB-1, as well as some SR shuttling proteins, and likely plays roles in pre-mRNA splicing and export of processed mRNA to the cytoplasm (10, 87). Sugiura et al. (76) have also shown that TLS/FUS binds proliferation-associated cytokine-inducible protein CIP29, a protein that interacts with URH49. DEAD-box RNA helicase UAP56 (and probably its paralog, URH49) binds the adaptor protein REF/Aly and recruits it to mRNA in the nucleus. Nuclear export factor 1 (NXF1/TAP) then binds REF/Aly, displacing UAP56, and targets the mRNA to the nuclear pore complex for export to the cytoplasm (39, 75, 76). The SR protein 9G8 also interacts with NXF1/TAP and promotes export of both intronless and spliced mRNAs from the nucleus (34). Significantly, Lindtner et al. (53) demonstrated that the L1 3′ UTR contains a novel sequence element that binds NXF1/TAP. Exploring connections between these proteins and L1 RNP transport from the nucleus, and possibly its reimport to the nucleus for retrointegration in the genome, should be possible with improved methods for visualizing L1 RNA in cells.
PAI-RBP1 was also found to interact with L1 ORF1p in an RNA-dependent manner. This protein was first isolated by its ability to bind PAI-1 mRNA and play an important role in its cyclic nucleotide-mediated stability (28). Significantly, VIG, the Drosophila homolog of PAI-RBP1, together with Fragile X protein dFMR1, was consistently copurified with the Drosophila RISC (9). A role for VIG in RNA interference has yet to be determined. Interestingly, disruption of the last exon of VIG, which is encoded within an intron of the DEAD-box RNA helicase Vasa gene, affected retrotransposon silencing in Drosophila. Unfortunately, this study did not ascertain if the effect was caused by Vasa or VIG (79).
Several lines of evidence suggest a direct role for RNA interference in L1 retrotransposon silencing. Small endogenous L1-related RNA products of a size consistent with siRNAs have been detected in human cell lines. Knockdown of Dicer 1, the RNase that processes siRNAs, caused an increase in the rate of retrotransposition of a tagged L1 in cell culture (84). Cotransfecting diced L1 siRNAs with an active L1 also decreased retrotransposition (72). Elevated transcription of murine L1s and IAP elements (an LTR retrotransposon) was observed in embryonic stem cells derived from Dicer-null mice (38). We have now presented further evidence for a connection between L1s and RNA interference by identifying L1 ORF1p in stress granules together with Ago2 and Fragile X protein FMRP, two known components of human RISC, as well as PAI-RBP1, a candidate RISC member.
Recently, stress granules have been linked with another mechanism that inhibits retrotransposition. Gallois-Montbrun et al. (20) reported that APOBEC3G relocalizes from PBs to SGs when cells are subjected to heat shock stress. Members of the APOBEC3 family of adenosine deaminases were originally defined by their ability to hypermutate reverse transcripts of human immunodeficiency virus RNA. Although only APOBEC3A, -B, -C, and -F have been reported to inhibit L1 retrotransposition (5, 66, 74), APOBEC3G inhibits L1-dependent retrotransposition of tagged Alu retroelements in cell culture (11). Interestingly, YB-1 (DBP-B) interacts with both APOBEC3F and APOBEC3G in an RNA-dependent manner and is found together with APOBEC3G in stress granules (20, 45). YB-1 also colocalizes with ORF1p in SGs and, along with the related protein DBP-A, was most frequently isolated from our two-hybrid interaction screens. A connection between YB-1 and SGs is not surprising since, among its many functions, the protein is thought to be involved in storing mRNAs in a translationally repressed state (reviewed in reference 73).
Retrotransposition has been a driving force of mammalian evolution over tens of millions of years. Approximately 100 L1 retrotransposons remain potentially active in humans and pose an ongoing threat to the integrity of the genome (7). Negative impacts are not limited to gene disruption by insertion or to recombination between dispersed repeat elements. L1 insertion can compromise gene function by introducing novel regulatory sequences having promoter/enhancer effects or that alter normal polyadenylation and splicing (reviewed in reference 27). Furthermore, recent studies have revealed that overexpressing L1 RNA and proteins may have deleterious consequences for cells, including induction of double-strand chromosomal breaks (22), apoptosis (3), and tumorigenesis (71). Faced with the potential damage L1s can cause, it is logical that the cell has evolved several mechanisms to hold retrotransposition in check. In addition to RNA interference and APOBEC-mediated suppression, promoter silencing by DNA methylation likely plays a major role. More than 90% of the methylated cytosines in the human genome occur in retrotransposons, and it has been proposed that DNA methylation evolved primarily as a defense mechanism against transposable elements (85).
We now speculate that cells have devised another retrotransposon defense: the sequestration of L1 RNPs in stress granules. By recognizing high levels of ORF1p as a stress, and targeting the protein and its bound RNA to SGs, the cell would reduce the number of L1 RNA molecules available for translation and nuclear import and thereby reduce retrotransposition. From SGs, ORF1p-bound RNA may be targeted to processing bodies for degradation. Furthermore, sequestering ORF1 protein in stress granules for degradation may prevent it from promiscuously binding non-L1 mRNAs, which might interfere with their proper trafficking or translation, or facilitate their retrotransposition into the genome as processed pseudogenes. This ORF1p sequestration would be most important in germ line precursor cells in which L1 protein and RNA have been detected in significantly higher levels than in somatic cells (6, 16).
On the other hand, stress granules may not be the end of the line for their resident mRNAs, which may escape degradation and return to the polysome fraction for reinitiation following stress recovery (41). In the absence of stress induction, we did not detect significant colocalization of ORF1p with either PABP or polyadenylated mRNA. This indicates that a large amount of non-L1 mRNA is present with ORF1p in cytoplasmic aggregates mainly when the cell is stressed. This influx of RNA to SGs likely increases the chance that L1 RNPs will capture non-L1 RNAs and, if released, possibly carry these to the nucleus for insertion in the genome. One might therefore predict that the rate of L1-mediated processed pseudogene creation would be higher in cells subjected to bouts of stress.
Furthermore, stress granules could play a role in the life cycle of L1s, rather than their death. For example, Beliakova-Bethell et al. (4) have shown that Ty3 LTR retrotransposons target yeast PBs where their RNA is sequestered away from translation and permitted to package in virus-like particles. Perhaps association with cytoplasmic RNA granules is involved in L1 RNP assembly and maturation in an analogous manner.
Supplementary Material
Acknowledgments
We greatly appreciate the reagents sent by the many researchers listed in the Materials and Methods section. We thank Eric Ostertag for critical reading of the manuscript. Special thanks to Gary Swain and the fabulous Morphology Core (Center Grant P30 DK50306), Division of Gastroenterology, University of Pennsylvania.
This work was supported by NIH/NIGMS grant 1R01GM45398-16 to H.H.K. and DOD/CDMRP grant BC051386 to J.L.G.
Footnotes
Published ahead of print on 11 June 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172:803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basame, S., P. Wai-lun Li, G. Howard, D. Branciforte, D. Keller, and S. L. Martin. 2005. Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition. J. Mol. Evol. 357:351-357. [DOI] [PubMed] [Google Scholar]
- 3.Belgnaoui, S. M., R. G. Gosden, O. J. Semmes, and A. Haoudi. 2006. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beliakova-Bethell, N., C. Beckam, T. H. Giddings, Jr., M. Winey, R. Parker, and S. Sandmeyer. 2006. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 12:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Branciforte, D., and S. L. Martin. 1994. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol. Cell. Biol. 14:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brouha, B., J. Schustak, R. M. Badge, S. Lutz-Prigge, A. H. Farley, J. V. Moran, and H. H. Kazazian, Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 100:5280-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buzdin, A., E. Gogvadze, E. Kovalskaya, P. Volchkov, S. Ustyugova, A. Illarionova, A. Fushan, T. Vinogradova, and E. Sverdlov. 2003. The human genome contains many types of chimeric retrogenes generated through in vivo RNA recombination. Nucleic Acids Res. 31:4385-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caudy, A. A., M. Myers, G. J. Hannon, and S. M. Hammond. 2002. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 16:2491-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chansky, H. A., M. Hu, D. D. Hickstein, and L. Yang. 2001. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 61:3586-3590. [PubMed] [Google Scholar]
- 11.Chiu, Y. L., H. E. Witkowska, S. C. Hall, M. Santiago, V. B. Soros, C. Esnault, T. Heidmann, and W. C. Greene. 2006. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl. Acad. Sci. USA 103:15588-15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cokol, M., R. Nair, and B. Rost. 2000. Finding nuclear localization signals. EMBO Rep. 1:411-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dang, Y., N. Kedersha, L. W. Low, D. Romo, M. Gorospe, K. Randal, P. Anderson, and J. O. Liu. 2006. Eukaryotic initiation factor 2α-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870-32878. [DOI] [PubMed] [Google Scholar]
- 14.Degracia, D. J., and B. R. Hu. 2006. Irreversible translation arrest in the reperfused brain. J. Cereb. Blood Flow Metab. 27:875-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duvernell, D. D., and B. J. Turner. 1998. Swimmer 1, a new low-copy-number LINE family in teleost genomes with sequence similarity to mammalian L1. Mol. Biol. Evol. 15:1791-1793. [DOI] [PubMed] [Google Scholar]
- 16.Ergün, S., C. Buschmann, J. Heukeshoven, K. Dammann, F. Schnieders, H. Lauke, F. Chalajour, N. Kilic, W. H. Stratling, and G. G. Schumann. 2004. Cell type-specific expression of LINE-1 open reading frames 1 and 2 in fetal and adult human tissues. J. Biol. Chem. 279:27753-27763. [DOI] [PubMed] [Google Scholar]
- 17.Esnault, C., J. Maestre, and T. Heidmann. 2000. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 24:363-367. [DOI] [PubMed] [Google Scholar]
- 18.Evdokimova, V. M., C. L. We, A. S. Sitikov, P. N. Simonenko, O. A. Lazarev, K. S. Vasilenko, V. A. Ustinov, J. W. Hershey, and L. P. Ovchinnikov. 1995. The major protein of messenger ribonucleoprotein particles in somatic cells is a member of the Y-box binding transcription factor family. J. Biol. Chem. 270:3186-3192. [DOI] [PubMed] [Google Scholar]
- 19.Fleckner, J., M. Zhang, J. Valcarcel, and M. R. Green. 1997. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 11:1864-1872. [DOI] [PubMed] [Google Scholar]
- 20.Gallois-Montbrun, S., B. Kramer, C. M. Swanson, H. Byers, S. Lynham, M. Ward, and M. H. Malim. 2006. The antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P-bodies and stress granules. J. Virol. 81:2165-2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gallouzi, I. E., C. M. Brennan, M. G. Stenberg, M. S. Swanson, A. Eversole, N. Maizels, and J. A. Steitz. 2000. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc. Natl. Acad. Sci. USA 97:3073-3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gasior, S. L., T. P. Wakeman, B. Xu, and P. L. Deininger. 2006. The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357:1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilbert, N., S. Lutz, T. A. Morrish, and J. V. Moran. 2005. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25:7780-7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilks, N., N. Kedersha, M. Ayodele, L. Shen, G. Stoecklin, L. M. Dember, and P. Anderson. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell 15:5383-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodier, J. L., E. M. Ostertag, K. A. Engleka, M. C. Seleme, and H. H. Kazazian, Jr. 2004. A potential role for the nucleolus in L1 retrotransposition. Hum. Mol. Genet. 13:1041-1048. [DOI] [PubMed] [Google Scholar]
- 26.Guil, S., J. C. Long, and J. F. Caceres. 2006. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol. Cell. Biol. 26:5744-5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han, J. S., and J. D. Boeke. 2005. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? Bioessays 27:775-784. [DOI] [PubMed] [Google Scholar]
- 28.Heaton, J. H., W. M. Dlakic, M. Dlakic, and T. D. Gelehrter. 2001. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the type-1 plasminogen activator inhibitor mRNA. J. Biol. Chem. 276:3341-3347. [DOI] [PubMed] [Google Scholar]
- 29.Hohjoh, H., and M. F. Singer. 1997. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J. Mol. Biol. 271:7-12. [DOI] [PubMed] [Google Scholar]
- 30.Holmes, S. E., M. F. Singer, and G. D. Swergold. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267:19765-19768. [PubMed] [Google Scholar]
- 31.Reference deleted.
- 32.Horton, P., K.-J. Park, T. Obayashi, and K. Nakai. 2006. Protein subcellular localization prediction with WoLF PSORT. Proc. 4th Ann. Asia Pacific Bioinformatics Conference APBC06, Taipei, p. 39-48.
- 33.Huang, Y., and J. A. Steitz. 2001. Splicing factors SRp20 and 9G8 promote the nucleocytoplasmic export of mRNA. Mol. Cell 7:899-905. [DOI] [PubMed] [Google Scholar]
- 34.Huang, Y., R. Gattoni, J. Stevenin, and J. A. Steitz. 2003. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell 11:837-843. [DOI] [PubMed] [Google Scholar]
- 35.International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 36.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Lu, T. Hai, H. P. Harding, X. Wang, D. Ron, D. R. Cavener, and R. C. Wek. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 24:1365-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jin, P., D. C. Zarnescu, S. Ceman, M. Nakamoto, J. Mowrey, T. A. Jongens, D. L. Nelson, K. Moses, and S. T. Warren. 2004. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat. Neurosci. 7:113-117. [DOI] [PubMed] [Google Scholar]
- 38.Kanellopoulou, C., S. A. Muljo, A. L. Kung, S. Ganesan, R. Drapkin, T. Jenuwein, D. M. Livingston, and K. Rajewsky. 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 19:489-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kapadia, F., A. Pryor, T. H. Chang, and L. F. Johnson. 2006. Nuclear localization of poly(A)+ mRNA following siRNA reduction of expression of the mammalian RNA helicases UAP56 and URH49. Gene 384:37-44. [DOI] [PubMed] [Google Scholar]
- 40.Kedersha, N. L., M. Gupta, W. Li, I. Miller, and P. Anderson. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kedersha, N., M. R. Cho, W. Li, P. W. Yacono, S. Chen, N. Gilks, D. E. Golan, and P. Anderson. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fritzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim, S. H., W. K. Dong, I. J. Weiler, and W. T. Greenough. 2006. Fragile X mental retardation protein shifts between polyribosomes and stress granules after neuronal injury by arsenite stress or in vivo hippocampal electrode insertion. J. Neurosci. 26:2413-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kohno, Y., Y. Matsuki, A. Tanimoto, H. Izumi, T. Uchiumi, K. Kohno, S. Shimajiri, and Y. Sasaguri. 2006. Expression of Y-box-binding protein dbpC/contrin, a potentially new cancer/testis antigen. Br. J. Cancer 94:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kozak, S. L., M. Marin, K. M. Rose, C. Bystrom, and D. Kabat. 2006. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol. Chem. 281:29105-29119. [DOI] [PubMed] [Google Scholar]
- 46.Kulpa, D. A., and J. V. Moran. 2006. cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 13:655-660. [DOI] [PubMed] [Google Scholar]
- 47.la Cour, T., L. Kiemer, A. Mølgaard, R. Gupta, K. Skriver, and S. Brunak. 2004. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 17:527-536. [DOI] [PubMed] [Google Scholar]
- 48.Leibold, D. M., G. D. Swergold, M. F. Singer, R. E. Thayer, B. A. Dombroski, and T. G. Fanning. 1990. Translation of LINE-1 DNA elements in vitro and in human cells. Proc. Natl. Acad. Sci. USA 87:6990-6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lemos, T. A., and J. Kobarg. 2006. CGI-55 interacts with nuclear proteins and co-localizes to p80-coilin positive-coiled bodies in the nucleus. Cell. Biochem. Biophys. 44:463-474. [DOI] [PubMed] [Google Scholar]
- 50.Leung, A. K., J. M. Calabrese, and P. A. Sharp. 2006. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA 103:18125-18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li, W., Y. Li, N. Kedersha, P. Anderson, M. Emara, K. M. Swiderek, G. T. Moreno, and M. A. Brinton. 2002. Cell proteins TIA-1 and TIAR interact with the 3′ stem-loop of the West Nile virus complementary minus-strand RNA and facilitate virus replication. J. Virol. 76:11989-12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin, S. S., M. H. Nymark-McMahon, L. Yieh, and S. B. Sandmeyer. 2001. Integrase mediates nuclear localization of Ty3. Mol. Cell. Biol. 21:7826-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lindtner, S., B. K. Felber, and J. Kjems. 2002. An element in the 3′ untranslated region of human LINE-1 retrotransposon mRNA binds NXF1(TAP) and can function as a nuclear export element. RNA 8:345-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu, J., M. A. Valencia-Sanchez, G. J. Hannon, and R. Parker. 2005. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 7:719-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu, J., F. V. Rivas, J. Wohlschlegel, J. R. Yates III, R. Parker, and G. J. Hannon. 2005. A role for the P-body component GW182 in microRNA function. Nat. Cell. Biol. 7:1261-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Luan, D. D., and T. H. Eickbush. 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol. Cell. Biol. 15:3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo, Y., and S. Li. 2007. Genome-wide analyses of retrogenes derived from the human box H/ACA snoRNAs. Nucleic Acids Res. 35:559-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin, S. L. 1991. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11:4804-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin, S. L., and D. Branciforte. 1993. Synchronous expression of LINE-1 RNA and protein in mouse embryonal carcinoma cells. Mol. Cell. Biol. 13:5383-5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin, S. L., J. Li, and J. A. Weisz. 2000. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1. J. Mol. Biol. 304:11-20. [DOI] [PubMed] [Google Scholar]
- 61.Martin, S. L., D. Branciforte, D. Keller, and D. L. Bain. 2003. Trimeric structure for an essential protein in L1 retrotransposition. Proc. Natl. Acad. Sci. USA 100:13815-13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin, S. L., M. Cruceanu, D. Branciforte, P. W.-L. Li, S. C. Kwok, R. S. Hodges, and M. C. Williams. 2005. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 348:549-561. [DOI] [PubMed] [Google Scholar]
- 62a.Mazroui, R., M. E. Huot, S. Tremblay, C. Filion, Y. Labelle, and E. W. Khandjian. 2002. Trapping of messenger RNA by Fragile X Mental Retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 11:3007-3017. [DOI] [PubMed] [Google Scholar]
- 63.McEwen, E., N. Kedersha, B. Song, D. Scheuner, N. Gilks, A. Han, J. J. Chen, R. Anderson, and R. J. Kaufman. 2005. Heme-regulated inhibitor kinase-mediated phosphorylation of eukaryotic translation initiation factor 2 inhibits translation, induces stress granule formation, and mediates survival upon arsenite exposure. J. Biol. Chem. 280:16925-16933. [DOI] [PubMed] [Google Scholar]
- 64.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 65.Mourelatos, Z., J. Dostie, S. Paushkin, A. Sharma, B. Charroux, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. miRNPs: a novel class of ribonucleoproteins containing numerous micro-RNAs. Genes Dev. 16:720-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muckenfuss, H., M. Hamdorf, U. Held, M. Perkovic, J. Lower, K. Cichutek, E. Flory, G. G. Schumann, and C. Munk. 2006. APOBEC3 proteins inhibit human LINE-1 retrotransposition. J. Biol. Chem. 281:22161-22172. [DOI] [PubMed] [Google Scholar]
- 67.Pillai, R. S., S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by Let-7 microRNA in human cells. Science 309:1573-1576. [DOI] [PubMed] [Google Scholar]
- 68.Pryor, A., L. Tung, Z. Yang, F. Kapadia, T.-H. Chang, and L. F. Johnson. 2004. Growth-regulated expression and G0-specific turnover of the mRNA that encodes URH49, a mammalian DExH/D box protein that is highly related to the mRNA export protein UAP56. Nucleic Acids Res. 32:1567-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rehwinkel, J., I. Behm-Ansmant, D. Gatfield, and E. Izaurralde. 2005. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA 11:1640-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sen, G. L., and H. M. Blau. 2005. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 7:633-636. [DOI] [PubMed] [Google Scholar]
- 71.Sinibaldi-Vallebona, P., P. Lavia, E. Garaci, and C. Spadafora. 2006. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosomes Cancer 45:1-10. [DOI] [PubMed] [Google Scholar]
- 72.Soifer, H. S., A. Zaragoza, M. Peyvan, M. A. Behlke, and J. J. Rossi. 2005. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 33:846-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sommerville, J., and M. Ladomery. 1996. Masking of mRNA by Y-box proteins. FASEB J. 10:435-443. [DOI] [PubMed] [Google Scholar]
- 74.Stenglein, M. D., and R. S. Harris. 2006. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 281:16837-16841. [DOI] [PubMed] [Google Scholar]
- 75.Strasser, K., and E. Hurt. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413:648-652. [DOI] [PubMed] [Google Scholar]
- 76.Sugiura, T., K. Sakurai, and Y. Nagano. 2006. Intracellular characterization of DDX39, a novel growth-associated RNA helicase. Exp. Cell. Res. 313:782-790. [DOI] [PubMed] [Google Scholar]
- 77.Teysset, L., V. D. Dang, M. K. Kim, and H. L. Levin. 2003. A long terminal repeat-containing retrotransposon of Schizosaccharomyces pombe expresses a Gag-like protein that assembles into virus-like particles which mediate reverse transcription. J. Virol. 77:5451-5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tourriere, H., K. Chebli, L. Zekri, B. Courseland, J. M. Blanchard, E. Bertrand, and J. Tazi. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823-831. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Vagin, V. V., M. S. Klenov, A. I. Kalmykova, A. D. Stolyarenko, R. N. Kotelnikov, and V. A. Gvozdev. 2004. The RNA interference proteins and Vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 1:54-58. [PubMed] [Google Scholar]
- 80.Vasudevan, S., and J. A. Steitz. 2007. AU-rich element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 128:1105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weber, M. J. 2006. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genet. 2:e205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wei, W., N. Gilbert, S. L. Ooi, J. F. Lawler, E. M. Ostertag, H. H. Kazazian, J. D. Boeke, and J. V. Moran. 2001. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 21:1429-1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Xiong, Y., W. D. Burke, J. L. Jakubczak, and T. H. Eickbush. 1988. Ribosomal DNA insertion elements R1Bm and R2Bm can transpose in a sequence specific manner to locations outside the 28S genes. Nucleic Acids Res. 16:10561-10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang, N., and H. H. Kazazian, Jr. 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13:763-771. [DOI] [PubMed] [Google Scholar]
- 85.Yoder, J. A., C. P. Walsh, and T. H. Bestor. 1997. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 13:335-339. [DOI] [PubMed] [Google Scholar]
- 86.Zhang, Z., P. M. Harrison, Y. Liu, and M. Gerstein. 2003. Millions of years of evolution preserved: a comprehensive catalog of the processed pseudogenes in the human genome. Genome Res. 13:2541-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zinszner, H., R. Albalat, and D. Ron. 1994. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 8:2513-2526. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.