Abstract
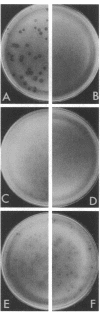
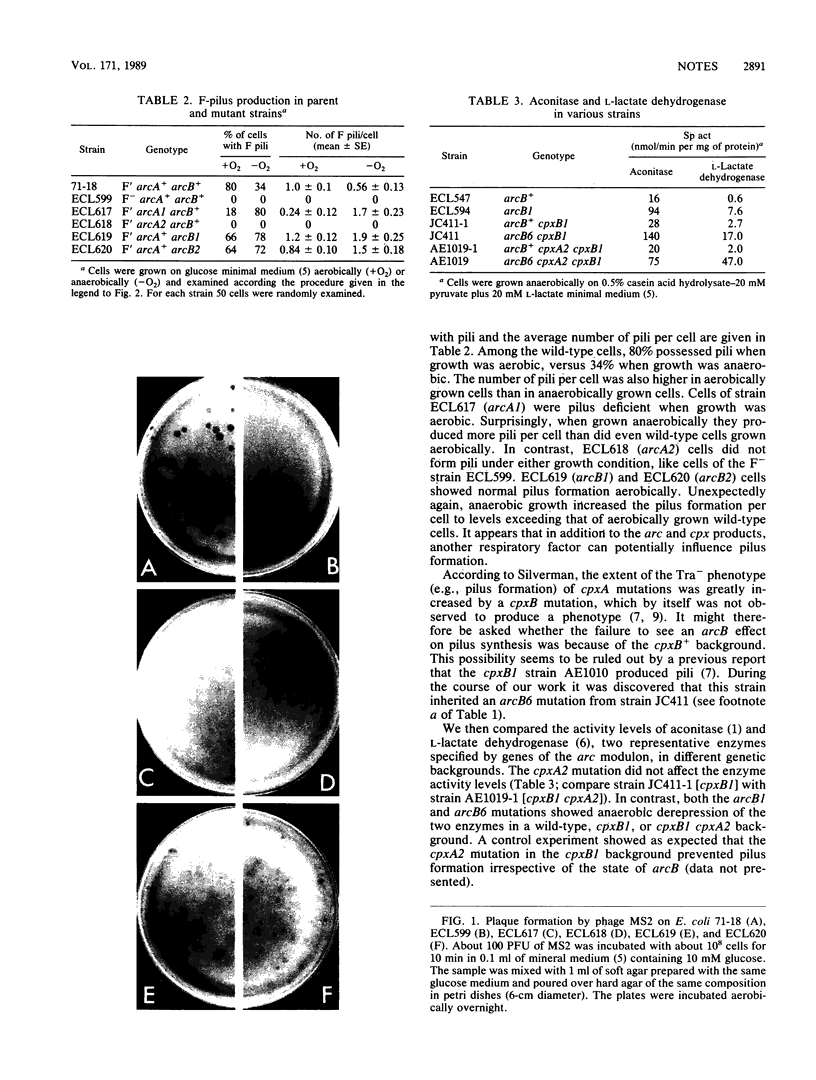
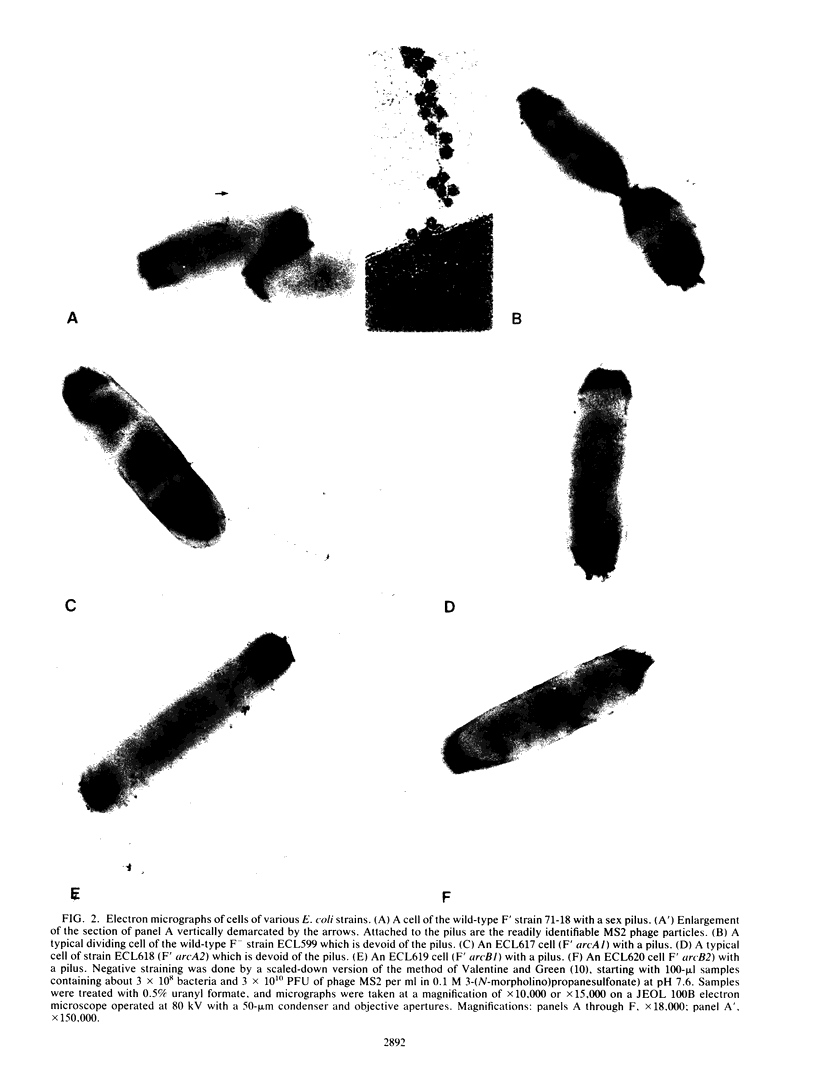
In Escherichia coli, mutations in arcA (dye) or arcB anaerobically derepress the synthesis of a multitude of enzymes of aerobic function, and mutations in arcA or cpxA impair F-pilus formation. It is thought that arcA encodes a promoter-recognizing protein, whereas arcB and cpxA encode sensor proteins which interact with the arcA product. In this study we found that anaerobic growth of a wild-type F' strain decreased the synthesis of both the enzymes and the pilus. Although the two arcA mutants examined were both anaerobically derepressed in the enzymes and impaired in aerobic pilus formation as expected, one mutant hyperproduced the pilus anaerobically. The two arcB mutants examined showed normal pilus formation when grown aerobically. When grown anaerobically they developed more pili than the wild-type strain did when grown aerobically. When a cpxA mutant was examined for synthesis of two aerobic enzymes, normal regulation was found. The available data suggest the following. The arcA product anaerobically represses certain genes of aerobic function and activates certain genes related to F function. It appears that the arcB product senses the redox or energy state; absence of the gene function shifts the arcA product to the nonrepressive form for enzyme synthesis for aerobic pathways. The cpxA product, on the other hand, senses the sexual state; absence of the gene function shifts the arcA product to the inactive form for F-pilus synthesis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Cameron D. C., Lin E. C. A second global regulator gene (arcB) mediating repression of enzymes in aerobic pathways of Escherichia coli. J Bacteriol. 1989 Feb;171(2):868–873. doi: 10.1128/jb.171.2.868-873.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J., Silverman P. Chromosomal mutations of Escherichia coli that alter expression of conjugative plasmid functions. Proc Natl Acad Sci U S A. 1980 Jan;77(1):513–517. doi: 10.1073/pnas.77.1.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronson C. W., Nixon B. T., Ausubel F. M. Conserved domains in bacterial regulatory proteins that respond to environmental stimuli. Cell. 1987 Jun 5;49(5):579–581. doi: 10.1016/0092-8674(87)90530-7. [DOI] [PubMed] [Google Scholar]
- Valentine R. C., Green N. M. Electron microscopy of an antibody-hapten complex. J Mol Biol. 1967 Aug 14;27(3):615–617. doi: 10.1016/0022-2836(67)90063-0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]