Abstract
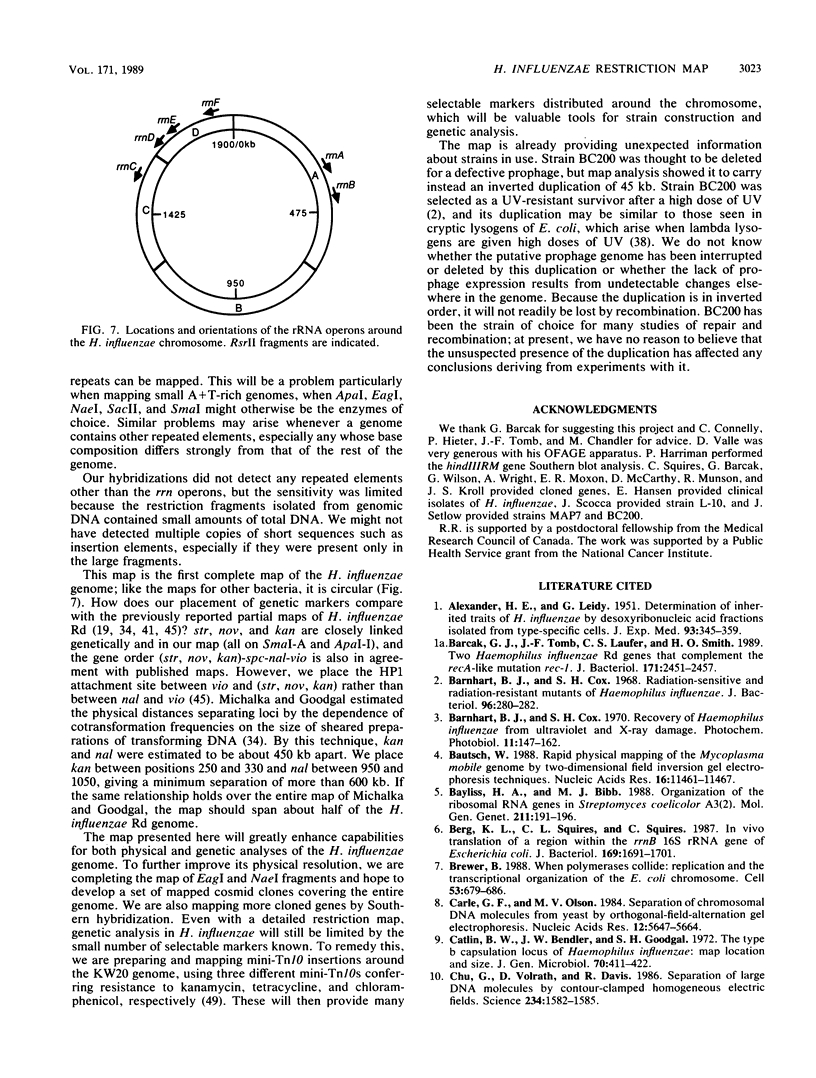
We present the first complete map of the Haemophilus influenzae genome, consisting of a detailed restriction map with a number of genetic loci. All of the ApaI, SmaI, and RsrII restriction sites (total of 45 sites) were mapped by Southern blot hybridization analysis of fragments separated by pulsed-field gel electrophoresis. Cloned genes were placed on the restriction map by Southern hybridization, and antibiotic resistance loci were also located by transformation with purified restriction fragments. The attachment site of the HP1 prophage was mapped. In addition, the number, locations, and orientations of the six rRNA operons in the H. influenzae chromosome were determined. The positions of conserved restriction sites in these rrn operons confirm that the direction of transcription is 16S to 23S, as in most other bacteria. The widely used strain BC200 appears to contain an unexpected 45-kilobase duplication.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ALEXANDER H. E., LEIDY G. Determination of inherited traits of H. influenzae by desoxyribonucleic acid fractions isolated from type-specific cells. J Exp Med. 1951 Apr 1;93(4):345–359. doi: 10.1084/jem.93.4.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcak G. J., Tomb J. F., Laufer C. S., Smith H. O. Two Haemophilus influenzae Rd genes that complement the recA-like mutation rec-1. J Bacteriol. 1989 May;171(5):2451–2457. doi: 10.1128/jb.171.5.2451-2457.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart B. J., Cox S. H. Radiation-sensitive and radiation-resistant mutants of Haemophilus influenzae. J Bacteriol. 1968 Jul;96(1):280–282. doi: 10.1128/jb.96.1.280-282.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart D. J., Cox S. H. Recovery of Haemophilus influenzae from ultraviolet and x-ray damage. Photochem Photobiol. 1970 Mar;11(3):147–162. doi: 10.1111/j.1751-1097.1970.tb05983.x. [DOI] [PubMed] [Google Scholar]
- Bautsch W. Rapid physical mapping of the Mycoplasma mobile genome by two-dimensional field inversion gel electrophoresis techniques. Nucleic Acids Res. 1988 Dec 23;16(24):11461–11467. doi: 10.1093/nar/16.24.11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylis H. A., Bibb M. J. Organisation of the ribosomal RNA genes in Streptomyces coelicolor A3(2). Mol Gen Genet. 1988 Feb;211(2):191–196. doi: 10.1007/BF00330593. [DOI] [PubMed] [Google Scholar]
- Berg K. L., Squires C. L., Squires C. In vivo translation of a region within the rrnB 16S rRNA gene of Escherichia coli. J Bacteriol. 1987 Apr;169(4):1691–1701. doi: 10.1128/jb.169.4.1691-1701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B. J. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988 Jun 3;53(5):679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984 Jul 25;12(14):5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catlin B. W., Bendler J. W., 3rd, Goodgal S. H. The type b capsulation locus of Haemophilus influenzae: map location and size. J Gen Microbiol. 1972 May;70(3):411–422. doi: 10.1099/00221287-70-3-411. [DOI] [PubMed] [Google Scholar]
- Chu G., Vollrath D., Davis R. W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986 Dec 19;234(4783):1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- Ellwood M., Nomura M. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J Bacteriol. 1982 Feb;149(2):458–468. doi: 10.1128/jb.149.2.458-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Gerardot C. J. Use of pulsed-field-gradient gel electrophoresis to construct a physical map of the Caulobacter crescentus genome. Gene. 1988 Sep 7;68(2):323–333. doi: 10.1016/0378-1119(88)90035-2. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Feingold J., Bellofatto V., Shapiro L., Amemiya K. Organization and nucleotide sequence analysis of an rRNA and tRNA gene cluster from Caulobacter crescentus. J Bacteriol. 1985 Jul;163(1):155–166. doi: 10.1128/jb.163.1.155-166.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzmaurice W. P., Scocca J. J. Restriction map and location of mutations on the genome of bacteriophage Hp1c1 of Haemophilus influenzae Rd. Gene. 1983 Sep;24(1):29–35. doi: 10.1016/0378-1119(83)90128-2. [DOI] [PubMed] [Google Scholar]
- Glaser G., Amikam D., Razin S. Physical mapping of the ribosomal RNA genes of Mycoplasma capricolum. Nucleic Acids Res. 1984 Mar 12;12(5):2421–2426. doi: 10.1093/nar/12.5.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Plaut A., Wright A. Haemophilus influenzae immunoglobulin A1 protease genes: cloning by plasmid integration-excision, comparative analyses, and localization of secretion determinants. J Bacteriol. 1987 Oct;169(10):4442–4450. doi: 10.1128/jb.169.10.4442-4450.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P. A., Patrick C. C., Hermanstorfer L., McCracken G. H., Jr, Hansen E. J. Conservation of epitopes in the oligosaccharide portion of the lipooligosaccharide of Haemophilus influenzae type b. Infect Immun. 1987 Mar;55(3):513–520. doi: 10.1128/iai.55.3.513-520.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herriott R. M., Meyer E. M., Vogt M. Defined nongrowth media for stage II development of competence in Haemophilus influenzae. J Bacteriol. 1970 Feb;101(2):517–524. doi: 10.1128/jb.101.2.517-524.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofman J. D., Lau R. H., Doolittle W. F. The number, physical organization and transcription of ribosomal RNA cistrons in an archaebacterium: Halobacterium halobium. Nucleic Acids Res. 1979 Nov 10;7(5):1321–1333. doi: 10.1093/nar/7.5.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huysmans E., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1986;14 (Suppl):r73–118. doi: 10.1093/nar/14.suppl.r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis E. D., Widom R. L., LaFauci G., Setoguchi Y., Richter I. R., Rudner R. Chromosomal organization of rRNA operons in Bacillus subtilis. Genetics. 1988 Nov;120(3):625–635. doi: 10.1093/genetics/120.3.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M., Burkard G., Bohnert H. J., Mubumbila M., Gordon K., Steinmetz A., Heiser D., Crouse E. J., Weil J. H. Transfer RNA genes associated with the 16S and 23S rRNA genes of Euglena chloroplast DNA. Biochem Biophys Res Commun. 1980 Jul 16;95(1):47–54. doi: 10.1016/0006-291x(80)90702-0. [DOI] [PubMed] [Google Scholar]
- Koch W., Edwards K., Kössel H. Sequencing of the 16S-23S spacer in a ribosomal RNA operon of Zea mays chloroplast DNA reveals two split tRNA genes. Cell. 1981 Jul;25(1):203–213. doi: 10.1016/0092-8674(81)90245-2. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Lee J. J., Smith H. O. Sizing of the Haemophilus influenzae Rd genome by pulsed-field agarose gel electrophoresis. J Bacteriol. 1988 Sep;170(9):4402–4405. doi: 10.1128/jb.170.9.4402-4405.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner A. F., Harvey S., Hill C. W. Mapping and spacer identification of rRNA operons of Salmonella typhimurium. J Bacteriol. 1984 Nov;160(2):682–686. doi: 10.1128/jb.160.2.682-686.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughney K., Lund E., Dahlberg J. E. tRNA genes are found between 16S and 23S rRNA genes in Bacillus subtilis. Nucleic Acids Res. 1982 Mar 11;10(5):1607–1624. doi: 10.1093/nar/10.5.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunnen K. D., Barsomian J. M., Camp R. R., Card C. O., Chen S. Z., Croft R., Looney M. C., Meda M. M., Moran L. S., Nwankwo D. O. Cloning type-II restriction and modification genes. Gene. 1988 Dec 25;74(1):25–32. doi: 10.1016/0378-1119(88)90242-9. [DOI] [PubMed] [Google Scholar]
- McCarthy D. Cloning of the rec-2 locus of Haemophilus influenzae. Gene. 1989 Jan 30;75(1):135–143. doi: 10.1016/0378-1119(89)90390-9. [DOI] [PubMed] [Google Scholar]
- McCarthy D., Cox S. S. rpe, a cis-acting element from the strA region of the Haemophilus influenzae chromosome that makes plasmid establishment independent of recombination. J Bacteriol. 1986 Oct;168(1):186–191. doi: 10.1128/jb.168.1.186-191.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalka J., Goodgal S. H. Genetic and physical map of the chromosome of Hemophilus influenzae. J Mol Biol. 1969 Oct 28;45(2):407–421. doi: 10.1016/0022-2836(69)90115-6. [DOI] [PubMed] [Google Scholar]
- Morgan E. A., Ikemura T., Nomura M. Identification of spacer tRNA genes in individual ribosomal RNA transcription units of Escherichia coli. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2710–2714. doi: 10.1073/pnas.74.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson R., Jr, Grass S. Purification, cloning, and sequence of outer membrane protein P1 of Haemophilus influenzae type b. Infect Immun. 1988 Sep;56(9):2235–2242. doi: 10.1128/iai.56.9.2235-2242.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Kroll J. S., Moxon E. R., Selander R. K. Clonal population structure of encapsulated Haemophilus influenzae. Infect Immun. 1988 Aug;56(8):1837–1845. doi: 10.1128/iai.56.8.1837-1845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redfield R. J., Campbell A. M. Structure of cryptic lambda prophages. J Mol Biol. 1987 Dec 5;198(3):393–404. doi: 10.1016/0022-2836(87)90289-0. [DOI] [PubMed] [Google Scholar]
- Roy P. H., Smith H. O. DNA methylases of Hemophilus influenzae Rd. I. Purification and properties. J Mol Biol. 1973 Dec 25;81(4):427–444. doi: 10.1016/0022-2836(73)90515-9. [DOI] [PubMed] [Google Scholar]
- Schwartz D. C., Cantor C. R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984 May;37(1):67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- Setlow J. K., Cabrera-Juárez E., Griffin K. Mechanism of acquisition of chromosomal markers by plasmids in Haemophilus influenzae. J Bacteriol. 1984 Nov;160(2):662–667. doi: 10.1128/jb.160.2.662-667.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L., Econome J. G., Schutt A., Klco S., Cantor C. R. A physical map of the Escherichia coli K12 genome. Science. 1987 Jun 12;236(4807):1448–1453. doi: 10.1126/science.3296194. [DOI] [PubMed] [Google Scholar]
- Stuy J. H. Transfer of genetic information within a colony of Haemophilus influenzae. J Bacteriol. 1985 Apr;162(1):1–4. doi: 10.1128/jb.162.1.1-4.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaiwa F., Sugiura M. Nucleotide sequence of the 16S - 23S spacer region in an rRNA gene cluster from tobacco chloroplast DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2665–2676. doi: 10.1093/nar/10.8.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Fitzmaurice W. P., Scocca J. J. Integration of the bacteriophage HP1c1 genome into the Haemophilus influenzae Rd chromosome in the lysogenic state. J Bacteriol. 1986 Jan;165(1):297–300. doi: 10.1128/jb.165.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Wenzel R., Herrmann R. Physical mapping of the Mycoplasma pneumoniae genome. Nucleic Acids Res. 1988 Sep 12;16(17):8323–8336. doi: 10.1093/nar/16.17.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]