Abstract
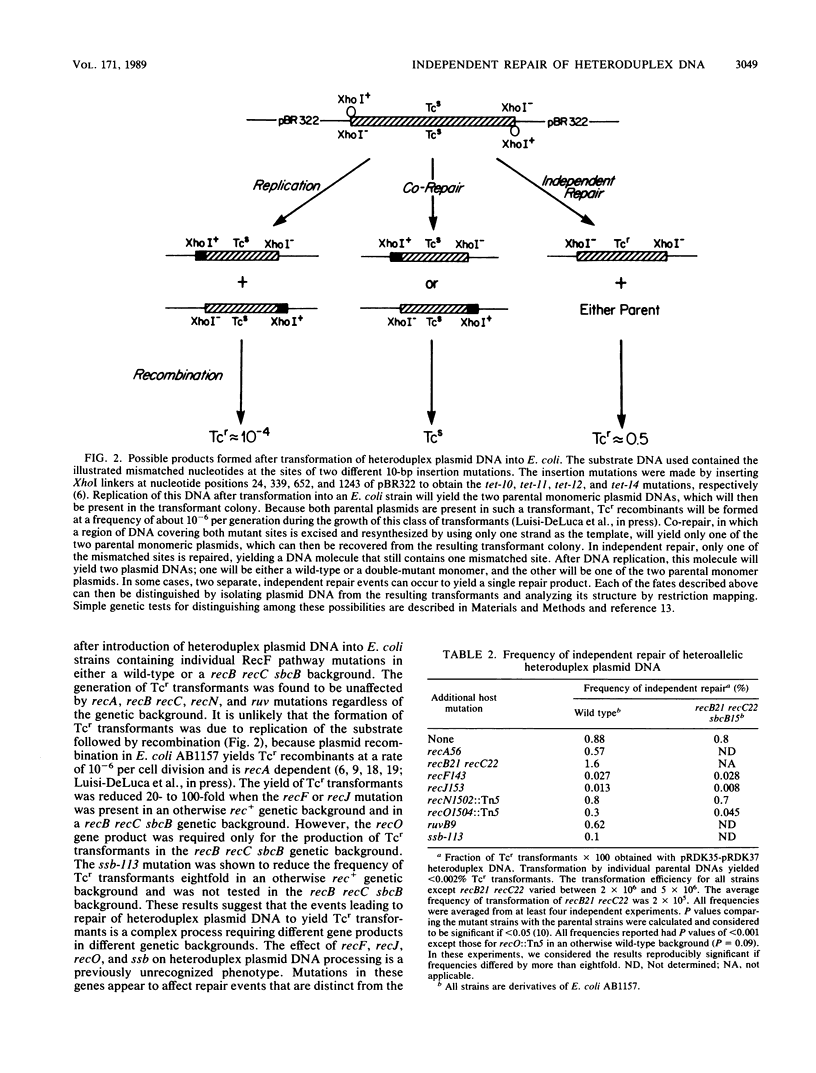
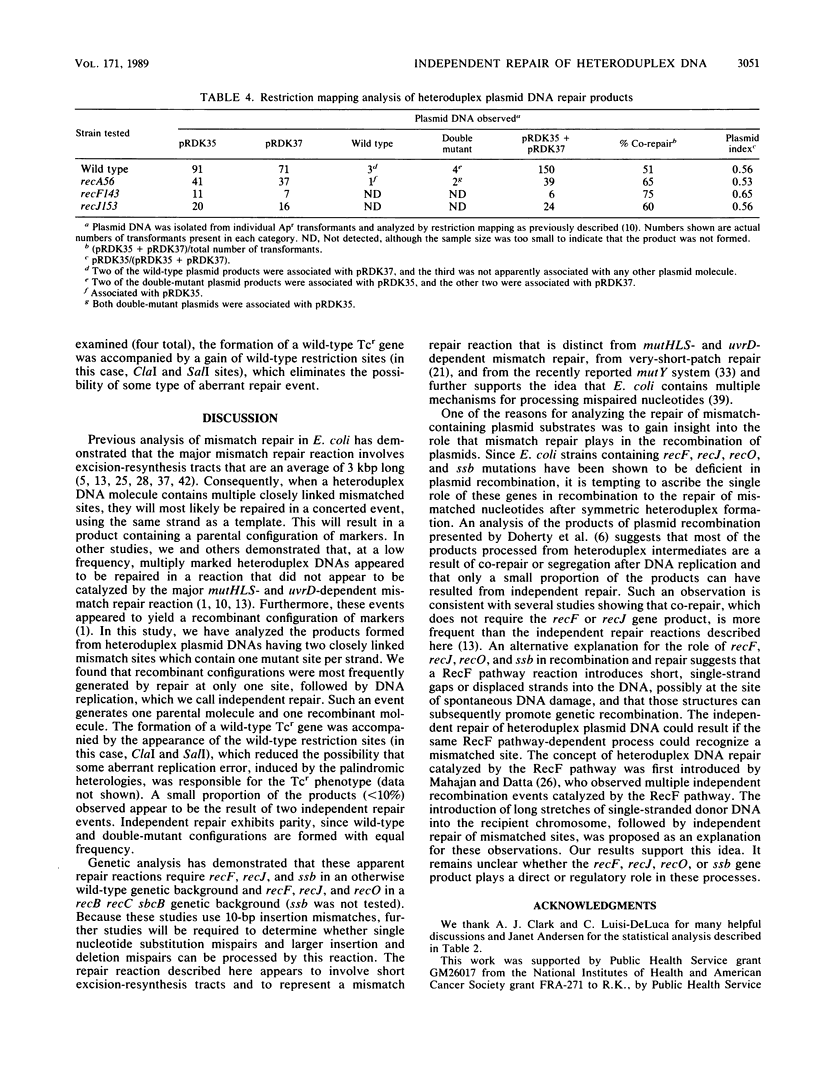
The independent repair of mismatched nucleotides present in heteroduplex DNA has been used to explain gene conversion and map expansion after general genetic recombination. We have constructed and purified heteroduplex plasmid DNAs that contain heteroallelic 10-base-pair insertion-deletion mismatches. These DNA substrates are similar in structure to the heteroduplex DNA intermediates that have been proposed to be produced during the genetic recombination of plasmids. These DNA substrates were transformed into wild-type and mutant Escherichia coli strains, and the fate of the heteroduplex DNA was determined by both restriction mapping and genetic tests. Independent repair events that yielded a wild-type Tetr gene were observed at a frequency of approximately 1% in both wild-type and recB recC sbcB mutant E. coli strains. The independent repair of small insertion-deletion-type mismatches separated by 1,243 base pairs was found to be reduced by recF, recJ, and ssb single mutations in an otherwise wild-type genetic background and reduced by recF, recJ, and recO mutations in a recB recC sbcB genetic background (the ssb mutation was not tested in the latter background). Independent repair of small insertion-deletion-type mismatched nucleotides that were as close as 312 nucleotides apart was observed. There was no apparent bias in favor of the insertion or deletion of mutant sequences.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abastado J. P., Cami B., Dinh T. H., Igolen J., Kourilsky P. Processing of complex heteroduplexes in Escherichia coli and Cos-1 monkey cells. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5792–5796. doi: 10.1073/pnas.81.18.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Volkert M. R., Margossian L. J. A role for recF in repair of UV damage to DNA. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):887–892. doi: 10.1101/sqb.1979.043.01.096. [DOI] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty M. J., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Physical and genetic analysis of the products of plasmid recombination in Escherichia coli. J Mol Biol. 1983 Jul 5;167(3):539–560. doi: 10.1016/s0022-2836(83)80097-7. [DOI] [PubMed] [Google Scholar]
- Dressler D., Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Feinstein S. I., Low K. B. Hyper-recombining recipient strains in bacterial conjugation. Genetics. 1986 May;113(1):13–33. doi: 10.1093/genetics/113.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fincham J. R., Holliday R. An explanation of fine structure map expansion in terms of excision repair. Mol Gen Genet. 1970;109(4):309–322. doi: 10.1007/BF00267701. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., Kolodner R. An Escherichia coli cell-free system that catalyzes the repair of symmetrically methylated heteroduplex DNA. Cold Spring Harb Symp Quant Biol. 1984;49:603–609. doi: 10.1101/sqb.1984.049.01.068. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., Kolodner R. Escherichia coli strains containing mutations in the structural gene for topoisomerase I are recombination deficient. J Bacteriol. 1984 Dec;160(3):1168–1170. doi: 10.1128/jb.160.3.1168-1170.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., Siegel E. C., Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986 Mar 20;188(2):147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Fox M. S. Some features of genetic recombination in procaryotes. Annu Rev Genet. 1978;12:47–68. doi: 10.1146/annurev.ge.12.120178.000403. [DOI] [PubMed] [Google Scholar]
- Herman G. E., Modrich P. Escherichia coli dam methylase. Physical and catalytic properties of the homogeneous enzyme. J Biol Chem. 1982 Mar 10;257(5):2605–2612. [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- James A. A., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Analysis of the effect of recombination-deficient mutations on plasmid recombination. J Mol Biol. 1982 Sep 25;160(3):411–430. doi: 10.1016/0022-2836(82)90305-9. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Fishel R. A., Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J Bacteriol. 1985 Sep;163(3):1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R. S., Su S. S., Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieb M. Specific mismatch correction in bacteriophage lambda crosses by very short patch repair. Mol Gen Genet. 1983;191(1):118–125. doi: 10.1007/BF00330898. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Benson F. E., Shurvinton C. E. Effect of ruv mutations on recombination and DNA repair in Escherichia coli K12. Mol Gen Genet. 1984;194(1-2):303–309. doi: 10.1007/BF00383532. [DOI] [PubMed] [Google Scholar]
- Lloyd R. G., Picksley S. M., Prescott C. Inducible expression of a gene specific to the RecF pathway for recombination in Escherichia coli K12. Mol Gen Genet. 1983;190(1):162–167. doi: 10.1007/BF00330340. [DOI] [PubMed] [Google Scholar]
- Lovett S. T., Clark A. J. Genetic analysis of the recJ gene of Escherichia coli K-12. J Bacteriol. 1984 Jan;157(1):190–196. doi: 10.1128/jb.157.1.190-196.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S. K., Datta A. R. Mechanisms of recombination by the RecBC and the RecF pathways following conjugation in Escherichia coli K12. Mol Gen Genet. 1979 Jan 16;169(1):67–78. doi: 10.1007/BF00267547. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Morrison D. A. Transformation in Escherichia coli: cryogenic preservation of competent cells. J Bacteriol. 1977 Oct;132(1):349–351. doi: 10.1128/jb.132.1.349-351.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama H., Nakayama K., Nakayama R., Irino N., Nakayama Y., Hanawalt P. C. Isolation and genetic characterization of a thymineless death-resistant mutant of Escherichia coli K12: identification of a new mutation (recQ1) that blocks the RecF recombination pathway. Mol Gen Genet. 1984;195(3):474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- Nevers P., Spatz H. C. Escherichia coli mutants uvr D and uvr E deficient in gene conversion of lambda-heteroduplexes. Mol Gen Genet. 1975 Aug 27;139(3):233–243. doi: 10.1007/BF00268974. [DOI] [PubMed] [Google Scholar]
- Nghiem Y., Cabrera M., Cupples C. G., Miller J. H. The mutY gene: a mutator locus in Escherichia coli that generates G.C----T.A transversions. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2709–2713. doi: 10.1073/pnas.85.8.2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pukkila P. J., Peterson J., Herman G., Modrich P., Meselson M. Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics. 1983 Aug;104(4):571–582. doi: 10.1093/genetics/104.4.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radding C. M. Genetic recombination: strand transfer and mismatch repair. Annu Rev Biochem. 1978;47:847–880. doi: 10.1146/annurev.bi.47.070178.004215. [DOI] [PubMed] [Google Scholar]
- Radman M., Wagner R. Mismatch repair in Escherichia coli. Annu Rev Genet. 1986;20:523–538. doi: 10.1146/annurev.ge.20.120186.002515. [DOI] [PubMed] [Google Scholar]
- Raposa S., Fox M. S. Some features of base pair mismatch and heterology repair in Escherichia coli. Genetics. 1987 Nov;117(3):381–390. doi: 10.1093/genetics/117.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper R. M. Escherichia coli mutator mutD5 is defective in the mutHLS pathway of DNA mismatch repair. Genetics. 1989 Feb;121(2):205–212. doi: 10.1093/genetics/121.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R. Homologous recombination in procaryotes. Microbiol Rev. 1988 Mar;52(1):1–28. doi: 10.1128/mr.52.1.1-28.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tait R. C., Rodriguez R. L., West R. W., Jr The rapid purification of T4 DNA ligase from a lambda T4 lig lysogen. J Biol Chem. 1980 Feb 10;255(3):813–815. [PubMed] [Google Scholar]
- White R. L., Fox M. S. On the molecular basis of high negative interference. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1544–1548. doi: 10.1073/pnas.71.4.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildenberg J., Meselson M. Mismatch repair in heteroduplex DNA. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2202–2206. doi: 10.1073/pnas.72.6.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]




