Abstract
Widespread and persistent marker expression is a prerequisite for many transgenic applications, including chimeric transplantation studies. Although existing transgenic tools for the clawed frog, Xenopus laevis, offer a number of promoters that drive widespread expression during embryonic stages, obtaining transgene expression through metamorphosis and into differentiated adult tissues has been difficult to achieve with this species. Here we report the application of the murine ROSA26 promoter in Xenopus. GFP is expressed in every transgenic tissue and cell type examined at post-metamorphic stages. Furthermore, transgenic ROSA26:GFP frogs develop normally, with no apparent differences in growth or morphology relative to wild-type frogs. ROSA26 transgenes may be used as a reliable marker for embryonic fate-mapping of adult structures in Xenopus laevis. Utility of this transgenic line is illustrated by its use in a chimeric grafting study that demonstrates the derivation of the adult bony jaw from embryonic cranial neural crest.
Keywords: bone, chimera, cranial neural crest, developmental biology, skull
Introduction
Fate maps of embryonic tissues and other cell populations have long been important for understanding the developmental and genetic underpinnings of adult morphologies (Landacre, 1921; Hörstadius, 1950; Sadaghiani & Thiébaud, 1987; Balaban et al. 1988; Clarke & Tickle, 1999). Along with the importance of fate maps for developmental studies is their utility for comparative biology. For example, detailed comparisons of fate maps among closely related species (e.g. vertebrates) inform our understanding of how alterations in the embryonic origin of a given structure may correlate with its morphological variation among taxa (Rudel & Sommer, 2003).
To yield useful results, any methodology used to generate a fate map must satisfy several criteria (reviewed in Kisseberth et al. 1999). For example, the method must reliably indicate a cell's lineage while minimizing the risk of false positives (i.e. ascribing an incorrect embryonic origin to a given structure) as well as false negatives (failure to detect the correct embryonic origin of a structure). A complication in metamorphic anurans, such as the well-studied model Xenopus laevis, is the protracted tadpole stage, which may last several months or longer (Trueb & Hanken, 1992). Any fate-mapping tool employed in this animal to assess the embryonic origin of adult-specific traits must last through this protracted life-history stage and into adulthood (Gross & Hanken, 2004).
Lineage tracers based on transgenes have the advantage of being intrinsic cellular markers; they do not dilute with time or age. However, in order to be useful for fate-mapping studies, transgenes must drive widespread and persistent expression. While promoters such as the Cytomegalovirus (CMV) promoter or the Xenopus elongation factor 1α (EF1α) gene promoter can be used to drive widespread expression during embryogenesis, the expression driven by these promoters is progressively restricted as tadpoles age and remains restricted in adult frogs (N. Marsh-Armstrong, unpublished data). Therefore, these promoters may be used as lineage tracers in young animals (e.g. De Robertis & Kuroda, 2004), but they are inadequate for assessing the derivation of adult traits. In the absence of a stable, long-term cell marker that persists through metamorphosis, experiments that explore the adult fate of embryonic cell populations in an amphibian model system have not been possible.
The fate of embryonic neural crest cells in the adult skull has historically received the most attention in avian models (Johnston et al. 1973; Le Lièvre, 1978; Noden, 1978; Couly et al. 1993; Köntges & Lumsden, 1996), but more recently it has also been addressed in the mouse (Chai et al. 2000; Morriss-Kay, 2001; Jiang et al. 2002; Matsuoka et al. 2005). Important differences have been reported in the neural crest contribution to skull bone between avian and mammalian models, raising the question as to the nature of the neural crest contribution in more primitive (basal) vertebrates (Hanken & Gross, 2005).
We aim to evaluate the neural crest contribution to skull bone in frogs as an extant model of a basal tetrapod. To this end, we have generated Xenopus laevis with an intrinsic cellular marker that is based on the ROSA26 gene. ROSA26 was identified originally as a ubiquitous marker in a retroviral gene-trapping screen in mouse embryonic stem cells (Friedrich & Soriano, 1991). The gene trap vector was later found to have integrated into and disrupted a ubiquitously expressed gene of unknown function (Zambrowicz et al. 1997). The promoter region for the ROSA gene was found to drive very widespread, if not ubiquitous, expression of reporter genes in transgenic mice (Kisseberth et al. 1999), albeit at levels of expression below those seen in the original ROSA26 mice (Zambrowicz et al. 1997). Mice carrying in their ROSA26 locus a conditional lacZ gene that expresses the reporter gene only after recombination due to Cre activity (Soriano, 1999) have become a common tool for genetic experiments. Recently, these mice have been used to study reporter expression in bone cell lineages (Lui et al. 2004), as well as the fate of neural crest cells in the skull (Jiang et al. 2002).
We use transgenic Xenopus laevis that express a green fluorescent protein (GFP) reporter gene under the control of the ROSA26 promoter, together with a chimeric grafting method, in order to obtain persistent GFP expression in explants of embryonic neural crest. The combination of embryonic grafting and a persistent and ubiquitous marker finally enables characterization of the adult, post-metamorphic fate of neural crest cells in an amphibian model system. These data will further our understanding of the nature of the contribution of osteogenic crest cells to the skull in a more comprehensive phylogenetic context.
Materials and methods
Incorporation of the ROSA26:GFP transgene into the Xenopus laevis genome via REMI
We used the technique of restriction enzyme-mediated integration (REMI) transgenesis in Xenopus laevis (Kroll & Amaya, 1996) to generate a transgenic line of frogs expressing the pR26-GFP plasmid (Kisseberth et al. 1999). Enhanced GFP (EGFP) is expressed under the control of a 0.8-kb fragment containing the ROSA26 promoter (plasmid kindly provided by Dr Eric Sandgren, University of Wisconsin, Madison). pR26-GFP, linearized with SalI, was purified with the GeneClean II kit (Qbiogene, Irvine, CA, USA) and used to make transgenic Xenopus laevis as previously described (Marsh-Armstrong et al. 1999).
Embryos and tadpoles made to express this transgene showed early and widespread GFP fluorescence (data not shown). A single female (F0) was raised to sexual maturity and outbred to wild-type animals. The F1 off-spring of these matings were used in the subsequent chimeric grafting experiments as well as analyses of the persistence of GFP expression in adult tissues. An F2 generation was produced from the colony of F1 individuals via in vitro fertilization using wild-type male sperm. Fertilizations were carried out using previously described methods (Sive et al. 2000).
Tissue processing and histology
Transgenic ROSA26:GFP and wild-type animals were fixed overnight at 4 °C in 3.7% PFA, pH 7.4. Specimens were rinsed several times for ∼1 h in non-sterile PBS solution. Following rinsing, several organs (heart, liver, colon, kidney) were removed from the transgenic and wild-type individuals. The tissues were rinsed again in PBS prior to immunohistological staining and processing.
Both the ROSA26:GFP and wild-type tissues were placed in a single 2-mL vial containing 5–10% normal goat serum (as a block) as well as a 1 : 2000 dilution of one or two of the following antibodies: Torrey Pines BioLabs rabbit polyclonal anti-GFP; AbCam rabbit polyclonal anti-GFP.
The vials were rocked on a nutator at 4 °C overnight (for at least 16 h) and then rinsed in multiple washes of PBS + 0.025% Triton X-100 at room temperature for 6–8 h. The tissues were placed back into a 2-mL vial containing a 1 : 500 dilution of goat anti-rabbit Alexa Fluor 488 (Molecular Probes, Eugene, OR, USA). Vials were again rocked at 4 °C overnight and then rinsed in multiple washes of PBS + 0.025% Triton X-100 at room temperature for 6–8 h. Once these tissues (heart, liver, colon and kidney) were analysed in whole-mount, they were cryosectioned and reanalysed in cross-section.
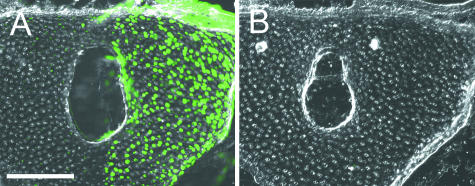
In addition to the tissues described above, various cranial tissues were assessed in cross-section for the presence of the GFP protein. Frontal cryosections were analysed for distribution of GFP (antibody stain, above) in a wild-type frog, a ROSA26:GFP transgenic frog and a mandibular stream cranial neural crest (CNC) grafted chimera (see Fig. 5). In our experiments, native GFP fluorescence did not persist strongly through the combination of protracted developmental times (i.e. after metamorphosis) and fixation employed to examine transgenic tissues. To determine the utility of antibody staining, we processed 16–20 µm-thick cryosections using an antibody staining protocol (see above) and compared them against neighbouring sections that were not processed immunohistochemically (Fig. 1). Positive label is present only on sections processed using the anti-GFP antibody staining protocol, which yielded significant amplification of the endogenous GFP signal (Fig. 1A) compared with untreated sections (Fig. 1B).
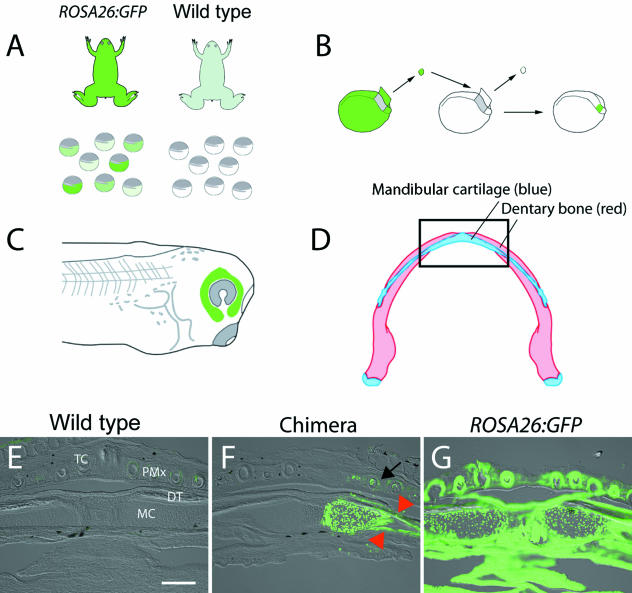
Fig 5.
Embryonic derivation of the adult lower jaw. We grafted premigratory cranial neural crest (CNC) from transgenic ROSA26:GFP donor embryos to wild-type host embryos at neurula stage, utilizing published fate maps (Sadaghiani & Thiébaud, 1987). In all experiments, eggs from both the ROSA26:GFP founder female and a wild-type female were fertilized in vitro with sperm from a wild-type male (A). ROSA26:GFP zygotes demonstrated variable intensity of GFP fluorescence; such variability is typical in animals with multiple transgene integration sites. Only the brightest, healthiest looking embryos were used as CNC explant donors (B). A tissue explant was removed from a transgenic ROSA26:GFP donor and replaced into a wild-type host. The explant was allowed to heal in place and was assayed the next morning to ensure proper development and migration of the CNC within the mandibular stream (C). Embryonic derivation of the adult mandibular cartilage and dentary bone in the lower jaw was assessed in chimeras that received a labelled graft of the mandibular stream of CNC. The jaw was sectioned in frontal plane (boxed region, D) and assessed for the presence of fluorescent label. GFP staining is absent in a wild-type specimen (E), whereas the entire jaw is labelled strongly in a ROSA26:GFP adult (G). Only the right (operated) side expresses GFP in a chimera that received the labelled graft (F). The left (control) side lacks the label, indicating that graft-derived cells did not cross to the contralateral side of the embryo or adult during development. Dense staining is evident within the extracellular matrix of the dentary bone (red arrowheads). Additional staining is present in the premaxillary bone of the upper jaw as well as portions of several tooth buds (black arrow). Abbreviations: DT, dentary bone; MC, mandibular cartilage; PMx, premaxillary bone; TC, tooth cusps. Scale bar (E–G), 250 µm.
Fig 1.
Use of a polyclonal anti-GFP antibody significantly amplifies GFP signal in cryosections of chimeric Xenopus. (A,B) Serial cross-sections through the rostral cartilage of an adult frog (NF stage 66, +2 months). Tissue processed using anti-GFP antibody (A; green dots) demonstrates robust staining for GFP. There is no detectable GFP expression in an adjacent section from the same individual (B; separated by ∼60 µm), which was processed without the antibody. In both sections, the right side received a grafted explant of cranial neural crest from a ROSA26:GFP transgenic donor; the left (ungrafted) side of the wild-type host provided an internal control. Scale bar, 200 µm.
Comparison of growth and morphology between transgenic ROSA26:GFP frogs and wild-type frogs
Transgenic ROSA26:GFP frogs and sibling wild-type frogs were divided among four groups: Group 1 – ROSA26:GFP, brightly fluorescing (n = 12); Group 2 – ROSA26:GFP, less brightly fluorescing (n = 31); Group 3 – wild-type set I (n = 12); Group 4 – wild type set II (n = 31). A sample of Group 1 individuals were killed and analysed for GFP expression in several tissues of the head and body (see below). All individual groups were reared in 4-L tanks with a separate supply of sterile, fresh 10% Holtfreter solution (Marine Biotech; Beverly, MA, USA) and fed ad libitum either live food (black worms) or frozen blood worms and frog brittle.
We observed no statistical difference in survival rate between the F1 ROSA26 and wild-type frogs (Pearson chi-square test, P = 0.238; Table 1). Because the frog transgenesis procedure with which the ROSA:GFP frog was generated is expected to increase the likelihood of developmental abnormalities, we carefully observed and characterized the normal growth and morphological development of this transgenic strain of frogs. All specimens were cleared and stained as whole-mounts according to standard procedures (Klymkowsky & Hanken, 1991).
Table 1.
Survivorship of ROSA26:GFP transgenic Xenopus laevis from 1 week post-fertilization (NF stage 42) to adult stage (NF 66), compared with wild-type frogs. There is no difference in survivorship between groups. Pearson chi-square test: 1 degree of freedom; χ2 = 1.397; P = 0.238
| Individual genetic background | Early tadpoles (NF stage 42) | Adults (NF stage 66 +2 months) | Total survival rate |
|---|---|---|---|
| Wild type | 44 | 15 | 34.1% |
| ROSA26:GFP (F1 generation) | 44 | 10 | 22.7% |
A set of adult ROSA26:GFP transgenic and wild-type individuals (n = 8 per group) were compared on six linear and one weight measurements (Table 2). All measures were tested for significance at the P = 0.05 level using a Student's t-test. Statistical analyses were carried out using Microsoft Excel (Version X) and SPSS (version 11.5).
Table 2.
Comparison of external growth measurements* reveals no difference between F1 transgenic frogs and wild-type frogs. Individual values indicate the mean measurement for each group ± standard deviation
| Individual genetic background | IOD | LCD | SVL | HL1 | HL2 | FL | W |
|---|---|---|---|---|---|---|---|
| Wild type (n = 8) | 4.48±0.32 | 7.33±0.46 | 24.36±2.94 | 26.67±2.63 | 32.46±3.66 | 12.13±1.01 | 1.74±0.70 |
| ROSA26:GFP (n = 8) | 4.82±0.39 | 7.77±0.57 | 24.67±3.73 | 29.22±4.11 | 34.61±5.23 | 12.46±1.64 | 1.81±0.95 |
| P-value | 0.08 | 0.12 | 0.86 | 0.16 | 0.36 | 0.64 | 0.85 |
IOD, interocular distance; LCD, lateral cranial distance; SVL, snout–vent length; HL1, hind limb length, measured from the longest toe to the hip when the limb is extended 90° from the long axis of the body; HL2, hind limb length, measured from the longest toe to the vent when the limb is extended 90° from the long axis of the body; FL, forelimb length; W, weight. Linear measurements are expressed in millimeters; weight is expressed in grams.
Southern hybridization
Genomic DNA was isolated from a wild-type and two transgenic members of the backcrossed F2 generation expressing either no detectable fluorescence compared with wild-type (−), or an individual expressing the strongest levels of GFP (+++). A sample of 20 µg of each was digested overnight with EcoRI. Genomic DNA was precipitated and run for 14 h on a 0.7% agarose gel at 20 V. Genomic DNA was then blotted to a Hybond nylon membrane for 12 h and processed for hybridization according to the method of Sambrook et al. (1989).
Probe was prepared by cutting out a ∼1-kb fragment of the GFP cassette from the R26R-GFP plasmid using AflII and BamHI. The probe was radiolabeled with 32P-dCTP using the Rediprime II kit (Amersham Pharmacia, Piscataway, NJ, USA). The probe was hybridized to the membrane overnight in UltraHyb buffer (Ambion, Austin, TX, USA), rinsed according to the manufacturer's instructions, and exposed for 4 days at −80 °C.
Embryonic tissue grafting
Embryo grafting was carried out at Nieuwkoop and Faber (NF) stage 14 (Nieuwkoop et al. 1994; Fig. 5B). Tissue explants of small portions of the CNC corresponding to the mandibular stream were grafted from ROSA26:GFP donor embryos into unlabelled wild-type hosts following standard procedures (Sadaghiani & Thiébaud, 1987; Gross & Hanken, 2005; Fig. 5B,C). All grafting was carried out on the right side; the left side of the developing head served as an internal negative control.
In each graft, a portion of the presumptive mandibular stream neural crest was first removed from an unlabelled wild-type host embryo and replaced with an equivalent-sized explant of the same region of the CNC from a labelled, donor transgenic embryo (Fig. 5B). Chimeric embryos were reared through metamorphosis, processed in cross-section, and analysed for the presence of donor-grafted GFP-positive cells in the adult bony jaw (Fig. 5F).
Image acquisition
Whole-mount specimens were placed in a Petri dish containing PBS + 0.025% Triton X-100 and photographed using a Leica MZ FLIII fluoroscope. Cryosectioned tissues were mounted with Fluoromount-G (Southern Biotech; Birmingham, AL, USA) and photographed using a Leica fluoroscope (model DMRE). High-resolution digital images were obtained with a 12-bit, black-and-white CCD camera (ORCA, Hamamatsu) using Openlab software (Improvision, UK). All tissues were stained and treated with identical antibody-staining protocols and photographic specifications.
All specimens were photographed in bright-field (cryosections were photographed using Nomarski optics) as well as fluorescence (B-filter, Leica) illumination. Black and white images were pseudo-coloured green using Openlab, merged with the light images and saved as TIFF files.
Intact adult specimens and cleared-and-stained skeletal preparations were photographed using an Automontage camera system (Syncroscopy USA, Frederick, MD, USA). All images were saved as TIFF files and compiled into figures using Adobe Photoshop 7.0.
Results
A new transgenic line of Xenopus laevis with widespread and persistent GFP expression
REMI transgenesis (Fig. 2A–C) was used to create embryos in which enhanced GFP (eGFP) expression was driven by a 0.8-kb proximal promoter fragment from the mouse ROSA26 gene (Kisseberth et al. 1999). These embryos had widespread transgene expression that, on average, was lower than that typically observed for the CMV promoter (data not shown). A single F0 ROSA26:GFP female was raised to adulthood. Upon breeding, this animal was found to carry multiple integrations of the transgene. There was variable GFP expression among the progeny: some had very weak fluorescence, whereas others had fluorescence comparable with that typically obtained with the CMV promoter (data not shown). We attribute the differences in expression to different integration sites – some allowing stronger expression than others – as well as a variable number of transgene integration sites.
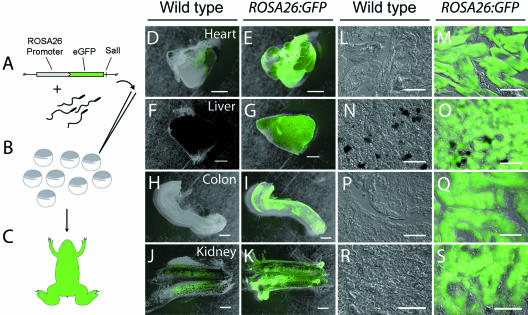
Fig 2.
Schematic depiction of REMI method of ROSA26:GFP transgenesis and anti-GFP staining of transgenic tissues. (A) A plasmid containing the ROSA26:GFP cassette is incubated with de-membranated wild-type sperm nuclei in a solution containing restriction enzyme (SalI). Resultant double-stranded breaks in the sperm nuclei genome allow for the introduction of the linearized plasmid. (B) Single nuclei are injected into unfertilized wild-type oocytes. (C) Animals carrying successful integration events and showing widespread GFP fluorescence are reared through metamorphosis. Offspring of a single founder female showing strong fluorescence were used as labelled, cranial neural crest (CNC) explant donors in all experiments. Several tissues were assayed for anti-GFP staining in whole-mounts and serial sections (D–S). In all tissues examined, the ROSA26 promoter drives GFP expression well into adulthood. Two wild-type organs processed for whole-mount immunohistochemistry, heart (D) and kidney (J), showed minor background staining that was much lower than the signal detected in comparable tissues from animals expressing the ROSA26:GFP transgene (E and K, respectively). The background staining was not evident in sections (L, R) when compared with ROSA26:GFP tissue (M, S). Scale bars: D–K, 1 mm; L–S, 100 µm.
The GFP reporter did not appear to affect either the survivability or the development of the founder female, further supporting the observation that GFP reporters are completely biosafe (Chalfie et al. 1994). We observed no developmental defects in the progeny of this founder (see Fig. 4, Table 2). As with other lines (Marsh-Armstrong et al. 1999), there appears to be no detectable change in either the pattern or the intensity of expression of the ROSA26:GFP transgene after passage through the germ line (data not shown). This enabled us to use the outbred offspring (F1) of this founder female (F0) in subsequent grafting experiments and analyses.
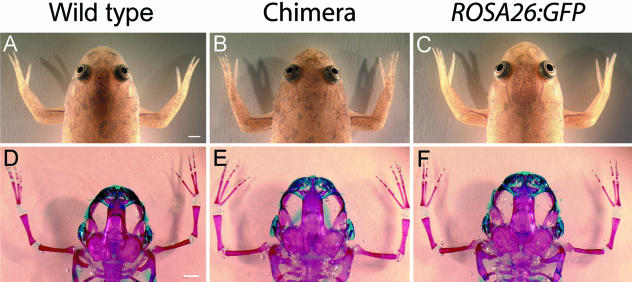
Fig 4.
External cranial morphology and skeletal development are indistinguishable among wild-type frogs (A,D), ROSA26:GFP transgenic frogs (C,F) and chimeric frogs (B,E). All animals were reared to at least 6 months of age; skeletal anatomy was assessed in cleared-and-stained whole-mounts (D–F). The three groups were indistinguishable from one another in terms of normal development and growth. The minor differences in cranial size among the three cleared-and-stained specimens depicted here are common even among populations of normal tadpoles reared under identical conditions but in different tanks. Scale bar, 2 mm.
GFP is detectable in a diverse array of organs and tissues throughout adulthood
To assess GFP expression in adult animals, we first analysed whole-mount, fixed tissues, including the heart, liver, colon and kidney. In each case, antibody staining revealed strong and uniform expression of the GFP protein (Fig. 2E,G,I,K). Minor fluorescence associated with the secondary antibody (AlexaFluor 488) was noted in the auricles of the heart (Fig. 2D) as well as restricted portions of the kidney (Fig. 2J). In both cases, this background fluorescence was negligible when compared with the intense fluorescence of ROSA26:GFP transgenic tissue, which had been processed with the identical staining and imaging protocols. Non-specific background staining that is occasionally present in whole-mount preparations is readily distinguished from specific, positive staining associated with the ROSA26:GFP construct.
All tissues were sectioned to determine the extent to which transgenic samples may be distinguished from wild-type samples at a fine microscopic level (Fig. 2L–S). In each comparison, the transgenic tissue showed strong and specific GFP signal whereas the wild-type tissue lacked any detectable signal. The slight background staining visible in some whole-mounts of wild-type tissues was not present in stained cryosections (Fig. 2L,R).
Some regions of the transgenic colon whole-mount appear to be unstained (Fig. 2I). We cannot explain the slightly lowered expression of GFP in this transgenic tissue preparation. We do note that in the corresponding sectioned preparation of the colon the signal is not diminished (Fig. 2Q), but instead it contrasts strongly with the complete and expected absence of signal in the wild-type section (Fig. 2P). It is unclear if the transgenic section shown (Fig. 2I) is demonstrative of all transgenic individuals, or only the brightest fluorescing individuals.
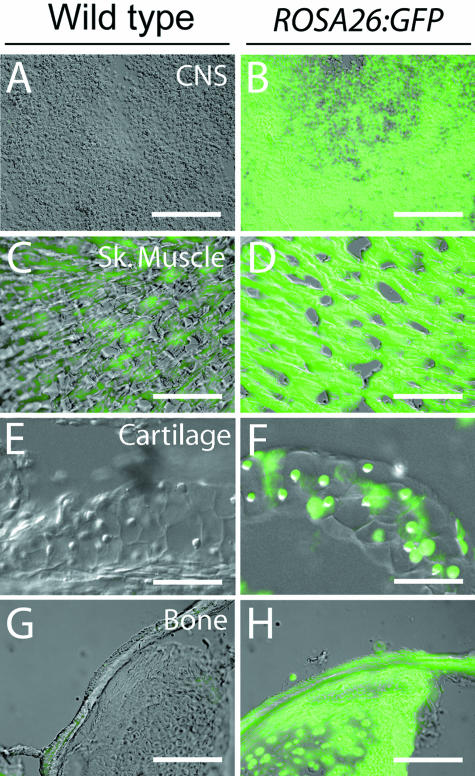
Comparison of reporter gene expression between transgenic and wild-type adult cranial tissues
Expression of the ROSA26:GFP reporter construct in cranial tissues was of special concern because of our interest in using this construct to assess direct contributions of cranial neural crest cells to the head of adult Xenopus laevis. Therefore, we surveyed cryosections of the prosencephalon, pterygoideus muscle, rostral cartilage and (ossified) pars articularis of the quadrate bone (Fig. 3).
Fig 3.
Antibody staining of adult cranial tissues. Frontal sections through the prosencephalon (A,B), pterygoideus muscle (C,D), rostral cartilage (E,F) and (ossified) pars articularis of the quadrate bone were immunostained to determine if the transgenic GFP label persists in these tissues. In all comparisons, anti-GFP staining in the ROSA26:GFP tissues (B,D,F,H) is clearly distinguishable when compared with wild-type tissues (A,C,E,G). Wild-type pterygoideus muscle showed minor background fluorescence (C) that is clearly distinguishable from the much more intense positive GFP staining of ROSA26:GFP muscle (D). CNS, central nervous system; Sk. muscle, skeletal muscle. Scale bar, 100 µm.
In every case, strong fluorescence was present in transgenic tissue (Fig. 3B,D,F,H) and generally absent in wild-type tissue (Fig. 3A,C,E,G). Some non-specific background staining is visible in wild-type skeletal muscle (Fig. 3C). As described above for whole-mounts, however, autofluorescence in wild-type tissue is minimal when compared with the fluorescence observed in transgenic tissue (Fig. 3D), and these two tissue types can be distinguished unequivocally according to staining intensity in chimeric grafting studies.
Transgenic, wild-type and chimeric individuals have indistinguishable cranial skeletal morphology and survival rates
Two important prerequisites for the use of ROSA26:GFP transgenic embryonic tissue in long-term fate-mapping studies are that transgenic and wild-type animals develop along identical timelines, and that the adult morphologies of these two groups are indistinguishable from one another. Therefore, we reared groups of transgenic and wild-type frogs under identical conditions and assessed their survivability and cranial morphologies (Fig. 4).
The survival rate of ROSA26:GFP transgenic frogs was slightly lower than that of wild-type frogs, but the difference between group means was not statistically significant (Table 1). Furthermore, there was no difference in adult body size or proportions between these two groups (Table 2). Both external and internal (skeletal) cranial morphology also appear to be identical among transgenic, wild-type and chimeric individuals (Fig. 4).
Use of the ROSA26:GFP transgenic frog for tracing adult derivatives of embryonic neural crest
To determine if the ROSA26:GFP transgene offers a reliable lineage tracer for long-term mapping studies, we grafted premigratory CNC cells that give rise to the mandibular stream from a transgenic embryo into an unlabelled wild-type host (Fig. 5A–C). The neural crest has long been recognized as the principal source of the chondrocranium in larval amphibians, including mandibular cartilages that constitute the larval lower jaw (Landacre, 1921; Sadaghiani & Thiébaud, 1987). Here, however, we were interested specifically in determining whether neural crest also contributes to bones of the lower jaw, which in anurans do not differentiate until metamorphosis and are fully formed only in adults.
Beginning 2 months following the end of metamorphosis (NF stage 66), we assessed the derivation of bone and cartilage of the anterior lower jaw in chimeric frogs that received ROSA26:GFP-labelled neural crest grafts (Fig. 5D). At this stage, anti-GFP staining strongly and uniformly labels all tissues in ROSA26:GFP frogs (Fig. 5G), whereas positive GFP label is absent in wild-type individuals (Fig. 5E). As expected, labelled chondrocytes were evident within the mandibular cartilage on the operated (right) side of the lower jaw (MC; Fig. 5F). However, label was also present within the calcified matrix of the dentary, one of two bones that form the anuran lower jaw (Fig. 5F, arrowheads). Additional label was present in the upper jaw, within the matrix of the premaxillary bone and in nearby tooth buds (Fig. 5F; arrow). GFP-positive tissues are absent on the un-operated (left) side of all chimeras (Fig. 5F).
Early survival and GFP expression among outbred F2 progeny
As a further test of the future utility of the ROSA26:GFP line of transgenic Xenopus, we outbred F1 females to wild-type sperm via in vitro fertilization and assessed early survival of the F2 generation. The transgenic embryos suffered a significantly higher mortality compared with wild-type fertilized eggs in the first 24 h post-fertilization (hpf; Table 3). Beyond 24 hpf, however, survivorship rates were comparable, and high, in both groups. More than 95% of embryos alive at 24 hpf remained alive at 48 hpf, and more than 90% of embryos alive at 48 hpf remained alive at 6 days post-fertilization (dpf).
Table 3.
Early survivorship of F2 ROSA26 embryos compared with wild-type embryos
| Individual genetic background | Total no. of fertilized eggs at day 0 | Embryos surviving between day 0 and 24 hpf* (NF stage 15/16) | Embryos surviving between 24 and 48 hpf (NF stage 33/34) | Tadpoles surviving between 48 hpf and 6 dpf (NF stage 44) |
|---|---|---|---|---|
| Wild type | 842 | 651 (77.3%) | 627 (96.3%) | 581 (92.7%) |
| F2 | 438 | 224 (51.1%) | 216 (96.4%) | 195 (90.3%) |
| Pearson chi-square results | χ2 = 91.26; P < 0.001 | χ2 = 0.006; P = 0.94 | χ2 = 1.25; P = 0.26 |
hpf, hours post-fertilization; dpf, days post-fertilization; NF, Nieuwkoop and Faber.
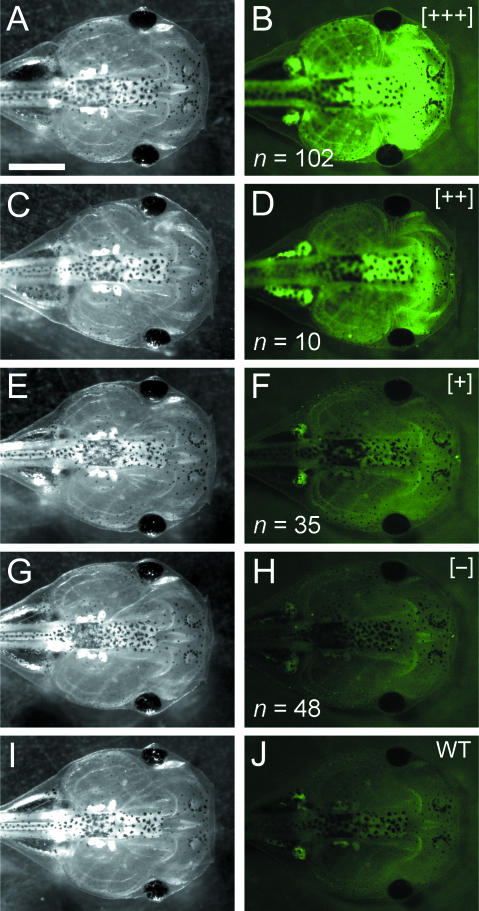
We also assessed variability in GFP expression across the outbred F2 generation. Endogenous fluorescence of live tadpoles was scored qualitatively at 6 dpf (approximately NF stage 44). Individuals were exposed to the same level of UV illumination and sorted among four groups (Fig. 6): (1) brightest fluorescence (+++), (2) intermediate fluorescence (++), (3) low-level fluorescence (+) and (4) no detectable fluorescence (−). Endogenous fluorescence in the ‘brightest’ and ‘intermediate’ individuals was very strong, especially compared with wild-type individuals (Fig. 6I,J). ‘Low-level’ individuals were only slightly brighter than those with no detectable fluorescence and wild-type tadpoles. Transplantation experiments that use the ROSA26:GFP transgenic line should target brightest individuals as tissue donors whenever possible.
Fig 6.
Variability of endogenous GFP fluorescence among live F2 individuals. Eggs from a single ROSA26 transgenic female (F1) were fertilized in vitro using wild-type sperm. Qualitative were recorded among the resulting F2 progeny viewed under identical conditions (constant time and intensity of exposure to UV illumination). These individuals were assigned among four groups according to qualitative measures of GFP fluorescence: brightest (B; +++), intermediate (D; ++), low (F; +) and undetectable (H; −). The last group is indistinguishable from wild-type individuals (J). There was no apparent difference in morphology among F2 individuals, nor between transgenic and wild-type individuals. The 3 : 1 ratio of GFP-expressing (B,D,F; n = 147) to non-GFP-expressing individuals (H; n = 48) suggests that the F1 female assayed in this experiment probably carries multiple copies of the ROSA26:GFP transgene. Scale bar, 1 mm.
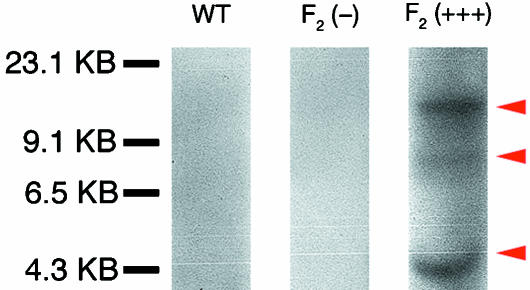
Because members of the F2 generation will be made available to our colleagues, we wished to determine whether the non-fluorescing (−) members of this generation carry the transgene. Towards this end, we performed a Southern hybridization of the genomic DNA from two representative members of this generation: (1) an individual not expressing fluorescence stronger than that of a wild-type individual (−), and (2) an individual displaying the strongest level of fluorescence both as an embryo and as a tadpole (+++). Our results show that the transgenic individual without apparent fluorescence [F2 (−); Fig. 6(G,H)] does not have any integrations of the transgene (Fig. 7). Furthermore, the brightly fluorescing individual [F2 (+++); Fig. 6(A,B)] has multiple copies of the transgene integrated into its genome (Fig. 7, arrowheads). At present we do not know the number of integration sites nor the number of copies per integration site. In light of these results, other investigators are encouraged to select the most brightly fluorescing individuals for long-term grafting projects.
Fig 7.
Southern blot analysis of members of the F2 generation of transgenic ROSA26 individuals. Genomic DNA from a wild-type individual (WT), an F2 individual expressing fluorescence at levels indistinguishable from wild-type (−), and an F2 individual expressing the brightest level of GFP fluorescence (+++) were analysed for the presence of transgene integration using Southern blot analysis. Both the wild-type individual and the transgenic F2-individual are shown not to carry the transgene. The F2 (+++) individual, however, carries multiple copies of the transgene (arrowheads). At present, it is not clear if fluorescence is correlated with the site of integration, number of copies at each integration site, or a combination of the two.
Discussion
ROSA26 promoter drives widespread reporter expression in adult tissues of Xenopus laevis
An essential requirement of any fate-mapping technique is that ‘labelled’ cells be both identifiable and distinguishable from surrounding ‘unlabelled’ cells in a chimeric environment (Kisseberth et al. 1999). Furthermore, the ideal fate-mapping technique should yield negligible background fluorescence. In every circumstance we considered, ROSA26:GFP transgenic tissues were much brighter and clearly distinguishable from wild-type tissues, especially when prepared as serial sections. Certain tissues may demonstrate endogenous autofluorescence due to a wide variety of causes unrelated to our experimental paradigm (e.g. flavoproteins and lipofuscins; Billington & Knight, 2001). In our study, this potential problem was confined principally to skeletal muscle, which displayed minor and non-specific background staining (autofluorescence) in sections. Use of ROSA26:GFP transgenic frogs for fate-mapping this particular tissue will require extra diligence to interpret staining patterns observed in chimeras correctly. Because there is variability in the levels of expression of different primary transgenic animals and the founder ROSA26:GFP female was not selected based on its expression level, it is likely that generating additional lines with the same ROSA26:GFP transgene will lead to lines with higher levels of expression and a higher signal-to-noise relative to endogenous autofluorescence.
We observed no statistically significant difference in survival rate to adulthood in ROSA26:GFP transgenic vs. wild-type frogs. The only survival difference we detected was confined to initial embryonic stages (0–24 hpf; Table 3). Large variability in survival rate is common even when breeding different wild-type animals. This early difference in survival rate might be a consequence of the sperm nuclei/REMI transgenesis procedure through which the ROSA26:GFP founder was created. Recently, alternative transgenesis procedures for Xenopus have been reported (Allen & Weeks, 2005; Pan et al. 2006), which may produce less deleterious effects than the sperm nuclei/REMI procedure. Future lines of ROSA26:GFP Xenopus may be generated with these novel methods as well. Finally, the presence of the transgene does not have any apparent effects on external or internal morphology (Fig. 4, Table 2).
The ROSA26:GFP transgene passes stably through the germ line
Numerous reporter constructs show stable passage through the germ line of Xenopus laevis (Marsh-Armstrong et al. 1999). We similarly find that the ROSA26:GFP cassette is capable of passing from F0 to F1 and F2 generations with no apparent change to the expression pattern or loss of fluorescence intensity of the reporter gene. The variability we observe in different animals is attributable to the fact that the one founder line for these studies had multiple integrations of the transgene. While the optimal line for future experiments would contain a single integration event of the transgene driving high expression of the reporter, we show here that the existing ROSA26:GFP line is suitable for grafting of labelled tissues, cellular progeny of which may be identified in both living and fixed specimens throughout all phases of the life history of Xenopus laevis.
Use of the ROSA26:GFP transgenic line as a novel tool for fate-mapping studies in Xenopus laevis
We previously reported derivation of the adult bony cranial vault of Xenopus laevis from embryonic neural crest using fluorescent dextran as a long-term marker (Gross & Hanken, 2005). Unfortunately, because the visible signal of fluorescent dextran diminishes over extended periods of time due to continued cell divisions and eventual autolysis of the tagged fluorophore, this method has limited applicability in studies that seek to map the embryonic derivation of adult-specific features.
Here we demonstrate that embryonic grafting, a standard technique in experimental embryology, may be combined with transgenic technology to produce chimeric embryos that express GFP in grafted tissues and their cellular progeny well into adulthood. For the first time, the embryonic origin of the bony lower jaw in adult Xenopus laevis is traced to the mandibular stream of cranial neural crest. Thus, along with the establishment of this line of transgenic frogs comes the ability to map the embryonic origin of a variety of adult tissues and organs, especially those that do not form until metamorphosis.
Use of the mouse ROSA26 promoter in frog transgenic studies
Based on widespread use of the ROSA26 locus in the mouse for genetic experiments that require ubiquitous transgene expression, we tested whether the murine ROSA26 promoter could drive similar expression in Xenopus. Such widespread and persistent transgene expression, which was not found in any of the previously available promoters for use in Xenopus, would enable, among other things, long-term fate-mapping studies in Xenopus that to date have been impossible.
Available sequence databases reveal no obvious homologue to the ROSA26 gene in Xenopus laevis or Silurana tropicalis, or any sequence in the Silurana tropicalis genome with high conservation to the murine ROSA26 promoter (data not shown). Interestingly, the murine promoter is still capable of driving widespread and persistent expression in Xenopus as it does in the mouse (Kisseberth et al. 1999). Thus, there appears to be functional conservation of this promoter in spite of the absence of obvious sequence conservation.
Unfortunately, little is known of the murine ROSA26 gene. It appears to encode an RNA transcribed in the opposite strand of another ubiquitously expressed gene, Thumpd3. Unlike the ROSA26 gene, Thumpd3 does appear to be conserved outside of mammals, including Xenopus. Thus, it is possible that the ROSA26 promoter works to regulate the expression of the Thumpd3 gene in both mice and frogs, although the Xenopus ROSA26 promoter cannot be identified with current algorithms. Alternatively, the ROSA26 promoter may function in frogs as it does in the mouse even without the equivalent gene in frogs.
Future applications of the ROSA26 promoter in Xenopus laevis
Whereas several promoters, including ROSA26, may drive widespread reporter expression in Xenopus embryos and very young tadpoles, ROSA26 is the only promoter of which we are aware that retains widespread expression in older tadpoles, through metamorphosis, and into adulthood. Thus, ROSA26 may be the promoter of choice for experiments that address these later ontogenetic stages and require widespread transgene expression. ROSA26 also may be a promoter of choice to be used in Xenopus transgenic studies that employ binary systems such as those based on Flp or Cre recombinases (Werdien et al. 2001; Ryffel et al. 2003; Gargioli & Slack, 2004) or in conjunction with ligand-inducible transgene expression systems in Xenopus (Das & Brown, 2004).
Conclusions
We have established a new line of transgenic frogs that express GFP under the control of the murine ROSA26 promoter. Reporter expression is present in unfertilized eggs and persists in every cell and tissue examined well into adulthood. This line of frogs offers a tractable tool for fate-mapping experiments because transgenic individuals grow along identical timelines as wild-type frogs with no apparent behavioral, developmental or morphological abnormalities. Incorporation of this transgenic line into ongoing studies of cranial development has further allowed us to apply a novel technique to solve a previously intractable problem, the fate of embryonic cells in the adult (post-metamorphic) head. We encourage use of this line of frogs for fate-mapping studies and as a step towards the introduction into Xenopus laevis of other genetic techniques (e.g. crosses utilizing site-specific recombinases) that until now have been limited mainly to mammalian model systems.
Acknowledgments
We thank Sarah Whitton for help with statistical analyses and Anne Everly for help with animal rearing and skeletal preparations. We are also grateful to Rahul Kanadia, Jennifer Mansfield, Douglas Kim and Maria Samson for help with Southern blot experiments. Eric Sandgren provided the pR26-GFP plasmid. Special thanks to Paul Trainor for help with GFP antibody staining. Funding was provided by the US National Science Foundation (EF-0334846; AmphibiaTree), The William F. Milton Fund (Harvard University) and a Goelet Summer Research Fellowship (Museum of Comparative Zoology).
References
- Allen BG, Weeks DL. Transgenic Xenopus laevis embryos can be generated using phiC31 integrase. Nat Meth. 2005;2:975–979. doi: 10.1038/nmeth814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban E, Teillet M-A, Le Douarin N. Application of the quail-chick chimera system to the study of brain development and behavior. Science. 1988;241:1339–1342. doi: 10.1126/science.3413496. [DOI] [PubMed] [Google Scholar]
- Billington N, Knight AW. Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence. Anal Biochem. 2001;291:175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, et al. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- Clarke JD, Tickle C. Fate maps old and new. Nat Cell Biol. 1999;1:E103–E109. doi: 10.1038/12105. [DOI] [PubMed] [Google Scholar]
- Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in quail-chick chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- Das B, Brown DD. Controlling transgene expression to study Xenopus laevis metamorphosis. Proc Natl Acad Sci USA. 2004;101:4839–4842. doi: 10.1073/pnas.0401011101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM, Kuroda H. Dorsal–ventral patterning and neural induction in Xenopus embryos. Annu Rev Cell Dev Biol. 2004;20:285–308. doi: 10.1146/annurev.cellbio.20.011403.154124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich G, Soriano P. Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev. 1991;5:1513–1523. doi: 10.1101/gad.5.9.1513. [DOI] [PubMed] [Google Scholar]
- Gargioli C, Slack JM. Cell lineage tracing during Xenopus tail regeneration. Development. 2004;131:2669–2679. doi: 10.1242/dev.01155. [DOI] [PubMed] [Google Scholar]
- Gross JB, Hanken J. Use of fluorescent dextran conjugates as a long-term marker of osteogenic neural crest in frogs. Dev Dynam. 2004;230:100–106. doi: 10.1002/dvdy.20036. [DOI] [PubMed] [Google Scholar]
- Gross JB, Hanken J. Cranial neural crest contributes to the bony skull vault in adult Xenopus laevis: insights from cell labeling studies. J Exp Zool (Mol Dev Evol) 2005;304B:169–176. doi: 10.1002/jez.b.21028. [DOI] [PubMed] [Google Scholar]
- Hanken J, Gross JB. Evolution of cranial development and the role of neural crest: insights from amphibians. J Anat. 2005;207:437–446. doi: 10.1111/j.1469-7580.2005.00481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörstadius S. The Neural Crest: its Properties and Derivatives in the Light of Experimental Research. London: Oxford University Press; 1950. [Google Scholar]
- Jiang X, Iseki S, Maxson RE, Sucov HM, Morriss-Kay GM. Tissue origins and interactions in the mammalian skull vault. Dev Biol. 2002;241:106–116. doi: 10.1006/dbio.2001.0487. [DOI] [PubMed] [Google Scholar]
- Johnston MC, Bhakdinaronk A, Reid YC. An expanded role of the neural crest in oral and pharyngeal development. In: Bosma JF, editor. Fourth Symposium on Oral Sensation and Perception: Development in the Fetus and Infant. Bethesda, MD: US Department of Health, Education, and Welfare; 1973. pp. 37–52. [PubMed] [Google Scholar]
- Kisseberth WC, Brettingen NT, Lohse JK, Sandgren EP. Ubiquitous expression of marker transgenes in mice and rats. Dev Biol. 1999;214:128–138. doi: 10.1006/dbio.1999.9417. [DOI] [PubMed] [Google Scholar]
- Klymkowsky MW, Hanken J. Whole-mount staining of Xenopus and other vertebrates. Meth Cell Biol. 1991;36:419–441. doi: 10.1016/s0091-679x(08)60290-3. [DOI] [PubMed] [Google Scholar]
- Köntges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kroll KL, Amaya E. Transgenic Xenopus embryos from sperm nuclear transplantations reveal FGF signaling requirements during gastrulation. Development. 1996;122:3173–3183. doi: 10.1242/dev.122.10.3173. [DOI] [PubMed] [Google Scholar]
- Landacre FL. The fate of the neural crest in the head of Urodeles. J Comp Neurol. 1921;33:1–43. [Google Scholar]
- Le Lièvre CS. Participation of neural crest-derived cells in the genesis of the skull in birds. J Embryol Exp Morph. 1978;47:17–37. [PubMed] [Google Scholar]
- Lui FL, Woitge HW, Braut A, et al. Expression and activity of osteoblast-targeted Cre recombinase transgenes in murine skeletal tissues. Int J Dev Biol. 2004;48:645–653. doi: 10.1387/ijdb.041816fl. [DOI] [PubMed] [Google Scholar]
- Marsh-Armstrong N, Huang H, Berry DL, Brown DD. Germ-line transmission of transgenes in Xenopus laevis. Proc Natl Acad Sci USA. 1999;96:14389–14393. doi: 10.1073/pnas.96.25.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T, Ahlberg PE, Kessaris N, et al. Neural crest origins of the neck and shoulder. Nature. 2005;436:347–355. doi: 10.1038/nature03837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morriss-Kay GM. Derivation of the mammalian skull vault. J Anat. 2001;199:143–151. doi: 10.1046/j.1469-7580.2001.19910143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J, Kirschner MW. Normal Table of Xenopus laevis (Daudin) New York: Garland Publishing Inc; 1994. [Google Scholar]
- Noden DM. The control of avian cephalic neural crest cytodifferentiation I. skeletal and connective tissues. Dev Biol. 1978;67:296–312. doi: 10.1016/0012-1606(78)90201-4. [DOI] [PubMed] [Google Scholar]
- Pan FC, Chen Y, Loeber J, Henningfeld K, Pieler T. I-SceImeganuclease-mediated transgenesis in Xenopus. Dev Dynam. 2006;235:247–252. doi: 10.1002/dvdy.20608. [DOI] [PubMed] [Google Scholar]
- Rudel D, Sommer RJ. The evolution of developmental mechanisms. Dev Biol. 2003;264:15–37. doi: 10.1016/s0012-1606(03)00353-1. [DOI] [PubMed] [Google Scholar]
- Ryffel GU, Werdien D, Turan G, Gerhards A, Goosses S, Senkel S. Tagging muscle cell lineages in development and tail regeneration using Cre recombinase in transgenic Xenopus. Nucleic Acids Res. 2003;31:e44. doi: 10.1093/nar/gng044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaghiani B, Thiébaud CH. Neural crest development in the Xenopus laevis embryo, studied by interspecific transplantation and scanning electron microscopy. Dev Biol. 1987;124:91–110. doi: 10.1016/0012-1606(87)90463-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sive H, Grainger RM, Harland RM. Early Development of Xenopus laevisA Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Trueb L, Hanken J. Skeletal development in Xenopus laevis (Anura: Pipidae) J Morph. 1992;214:1–41. doi: 10.1002/jmor.1052140102. [DOI] [PubMed] [Google Scholar]
- Werdien D, Peiler G, Ryffel GU. FLP and Cre recombinase function in Xenopus embryos. Nucleic Acids Res. 2001;29:E53–E53. doi: 10.1093/nar/29.11.e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]