Abstract
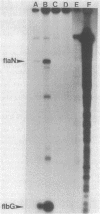
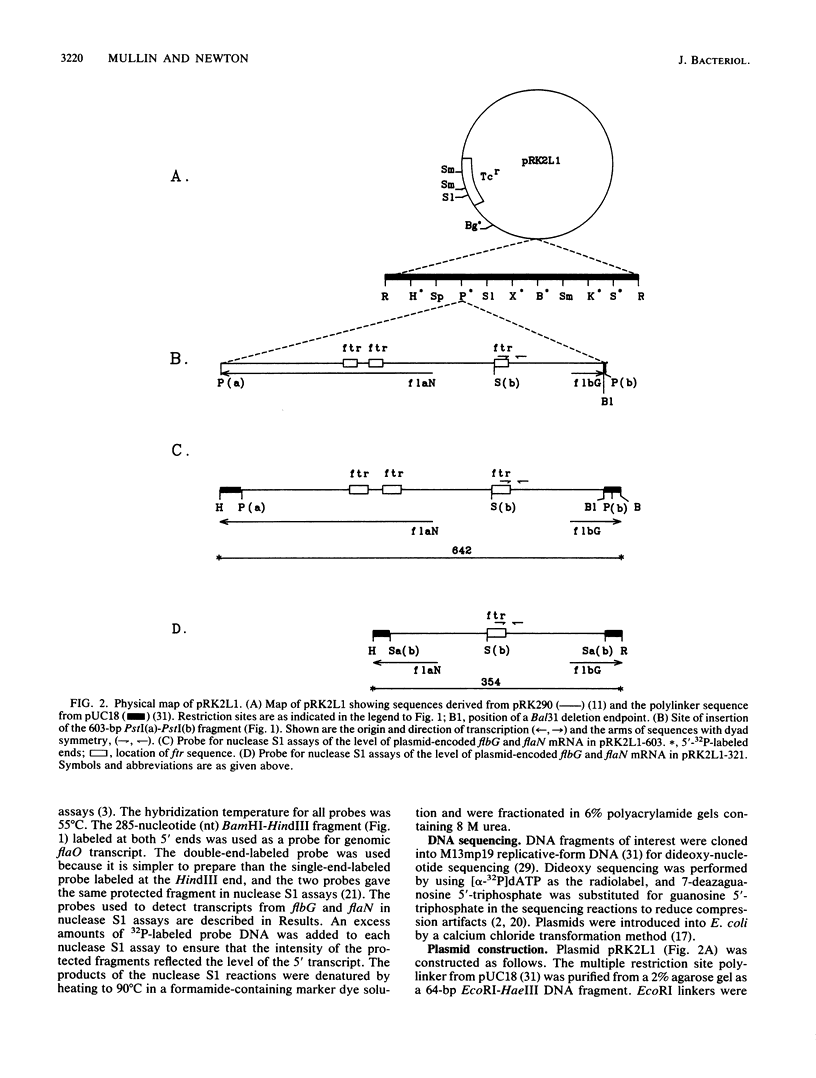
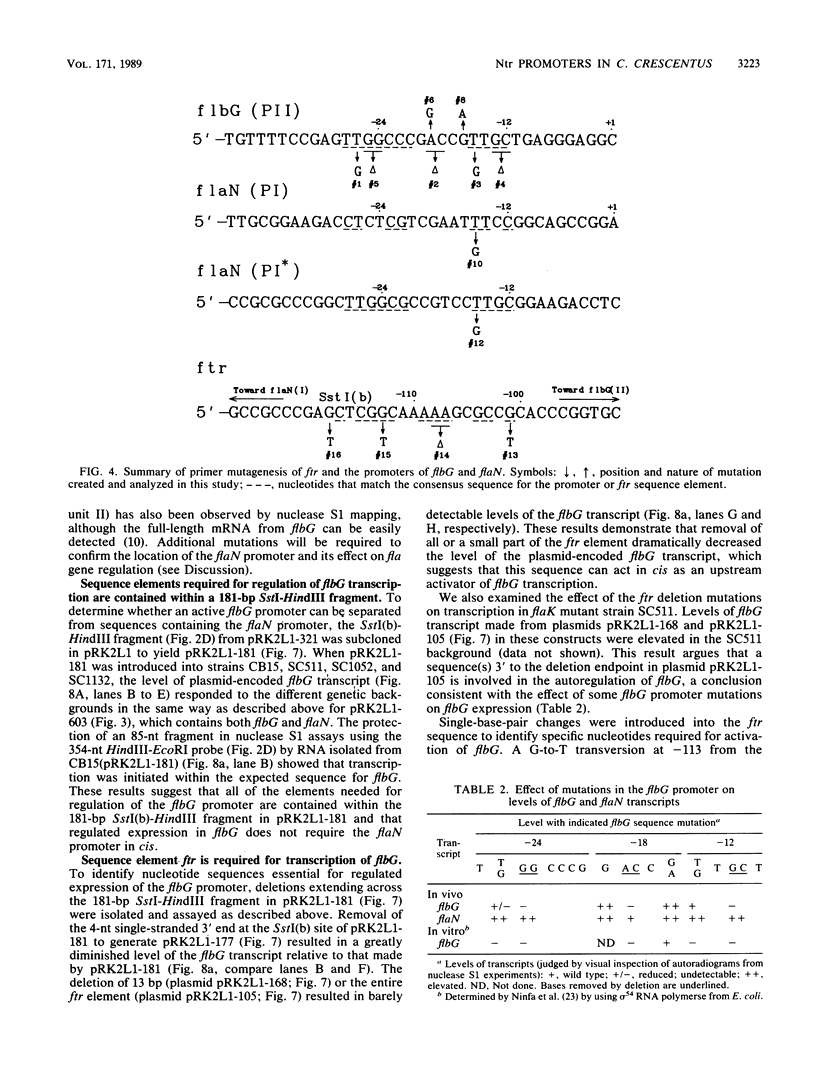
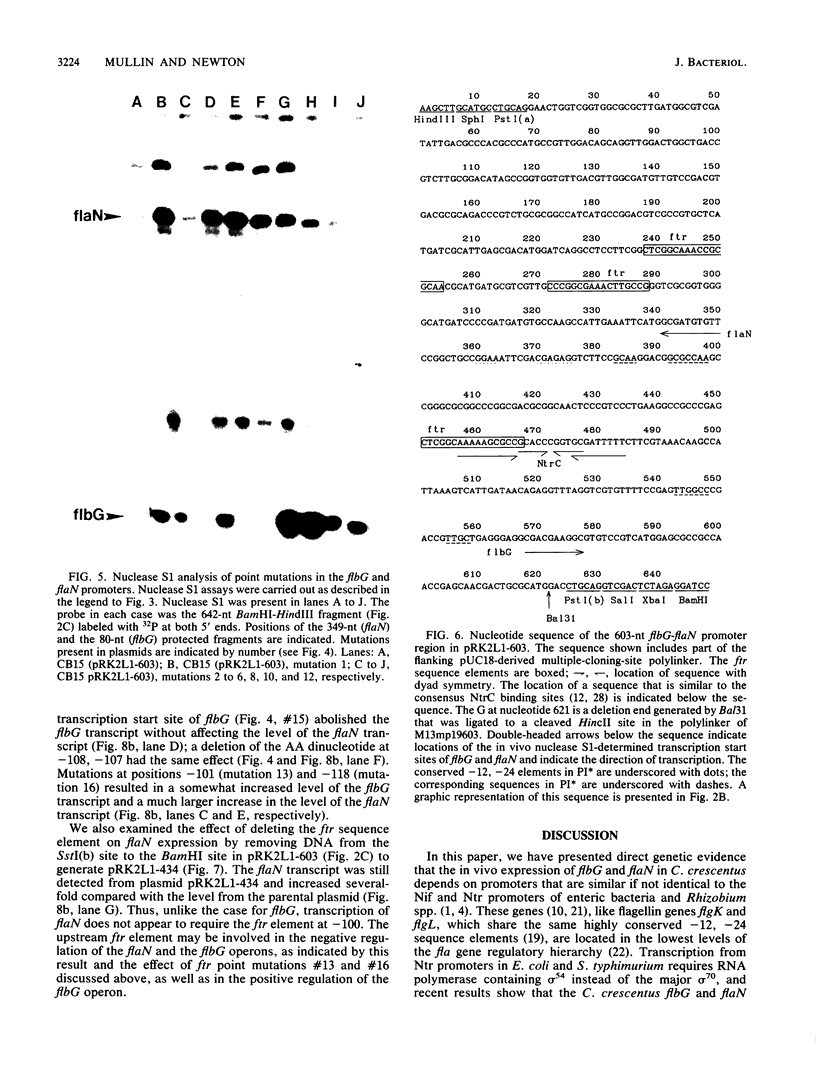
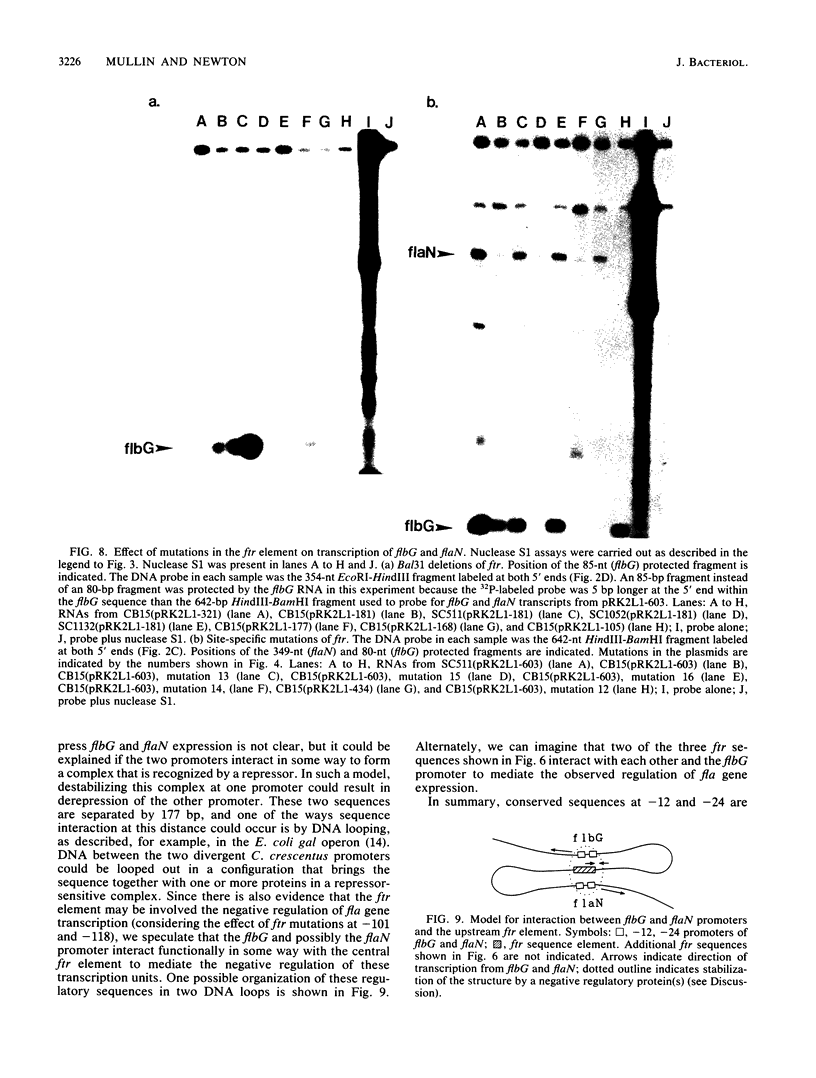
The flbG (hook operon or transcription unit II) and flaN (transcription unit I) operons of Caulobacter crescentus have a -12, -24 nucleotide sequence motif that is very similar to those of the Nif and Ntr promoters of enteric bacteria and Rhizobium spp. and a conserved ftr (flagellar gene transcription regulation) sequence, previously designated II-1 (D. A. Mullin, S. A. Minnich, L.-S. Chen, and A. Newton, J. Mol. Biol. 195:939-943, 1987) at approximately -100. We have used site-directed mutagenesis to examine the role of these sequences in the transcriptional regulation of these periodically expressed flagellar genes. Mutations in the flbG promoter that removed the conserved GC at -12, -13, the GG at -24, -25, or an AC base pair at -18, -19 in the nonconserved sequence between the -12, -24 elements completely eliminated detectable transcription. Mutations at other positions resulted in either a slight decrease (position 26), no change (position 15), or an elevated level (position -16 or -19) of the flbG transcript. By contrast, most of these flbG promoter mutations resulted in greatly elevated levels of transcription from the opposing flaN operon. Similar experiments were used to confirm the location of the flaN promoter to a -12, -24 Nif and Ntr sequence motif. Deletion of all or part of the ftr element or point mutations in the sequence drastically reduced the level of flbG transcript and resulted in increased levels of the flaN transcript. Thus, the conserved sequences at -12 and -24 in flbG and flaN are required for transcription of these genes in vivo, and the ftr element is required for transcription of flbG. This analysis also suggested that the ftr sequence and sequences in the flbG promoter are required for the negative autoregulation of the flbG and flaN operons. We speculate that the flbG and flaN promoters and the ftr element interact in some way to mediate the negative control of these divergent transcription units.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ausubel F. M. Regulation of nitrogen fixation genes. Cell. 1984 May;37(1):5–6. doi: 10.1016/0092-8674(84)90294-0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Beynon J., Cannon M., Buchanan-Wollaston V., Cannon F. The nif promoters of Klebsiella pneumoniae have a characteristic primary structure. Cell. 1983 Sep;34(2):665–671. doi: 10.1016/0092-8674(83)90399-9. [DOI] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brown S. E., Ausubel F. M. Mutations affecting regulation of the Klebsiella pneumoniae nifH (nitrogenase reductase) promotor. J Bacteriol. 1984 Jan;157(1):143–147. doi: 10.1128/jb.157.1.143-147.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M. Deletion analysis of the Klebsiella pneumoniae nitrogenase promoter: importance of spacing between conserved sequences around positions -12 and -24 for activation by the nifA and ntrC (glnG) products. J Bacteriol. 1986 May;166(2):545–551. doi: 10.1128/jb.166.2.545-551.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M., Khan H., Dixon R. Site-directed mutagenesis of the Klebsiella pneumoniae nifL and nifH promoters and in vivo analysis of promoter activity. Nucleic Acids Res. 1985 Nov 11;13(21):7621–7638. doi: 10.1093/nar/13.21.7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- Chen L. S., Mullin D., Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci U S A. 1986 May;83(9):2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo L., Martin K. J., Schleif R. Alternative DNA loops regulate the arabinose operon in Escherichia coli. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5444–5448. doi: 10.1073/pnas.85.15.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lagenaur C., Agabian N. Caulobacter flagellar organelle: synthesis, compartmentation, and assembly. J Bacteriol. 1978 Sep;135(3):1062–1069. doi: 10.1128/jb.135.3.1062-1069.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M., Cohen S. N. Transformation of Salmonella typhimurium by plasmid deoxyribonucleic acid. J Bacteriol. 1974 Sep;119(3):1072–1074. doi: 10.1128/jb.119.3.1072-1074.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewy Z. G., Bryan R. A., Reuter S. H., Shapiro L. Control of synthesis and positioning of a Caulobacter crescentus flagellar protein. Genes Dev. 1987 Aug;1(6):626–635. doi: 10.1101/gad.1.6.626. [DOI] [PubMed] [Google Scholar]
- Minnich S. A., Newton A. Promoter mapping and cell cycle regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin D., Minnich S., Chen L. S., Newton A. A set of positively regulated flagellar gene promoters in Caulobacter crescentus with sequence homology to the nif gene promoters of Klebsiella pneumoniae. J Mol Biol. 1987 Jun 20;195(4):939–943. doi: 10.1016/0022-2836(87)90497-9. [DOI] [PubMed] [Google Scholar]
- Ninfa A. J., Mullin D. A., Ramakrishnan G., Newton A. Escherichia coli sigma 54 RNA polymerase recognizes Caulobacter crescentus flbG and flaN flagellar gene promoters in vitro. J Bacteriol. 1989 Jan;171(1):383–391. doi: 10.1128/jb.171.1.383-391.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Swanson E., Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985 Nov 5;186(1):107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- Ohta N., Swanson E., Ely B., Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984 Jun;158(3):897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow D. W., Xiong Y., Gu Q., Shen S. C. Mutational analysis of the Klebsiella pneumoniae nitrogenase promoter: sequences essential for positive control by nifA and ntrC (glnG) products. J Bacteriol. 1985 Mar;161(3):868–874. doi: 10.1128/jb.161.3.868-874.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell. 1986 Jun 20;45(6):785–792. doi: 10.1016/0092-8674(86)90553-2. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Regulation of periodic protein synthesis in the cell cycle: control of initiation and termination of flagellar gene expression. Cell. 1981 Apr;24(1):49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]