Abstract
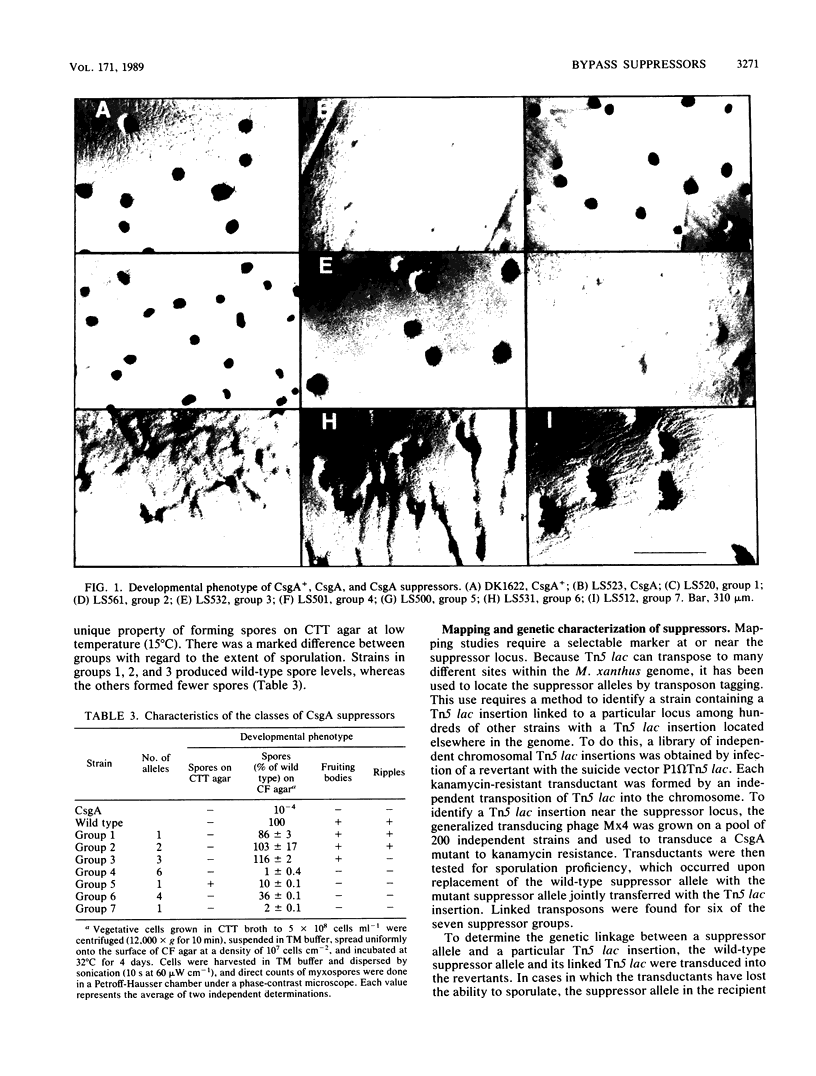
The csgA mutations of Myxococcus xanthus (formerly known as spoC) inhibit sporulation as well as rippling, which involves ridges of cells moving in waves. Sporulating revertants of CsgA cells were isolated by direct selection, since spores are much more resistant to heat and ultrasonic treatment than are vegetative cells. The revertants fell into seven groups on the basis of phenotype and the chromosomal location of the suppressor alleles. Group 1 contained one allele that was a back mutation of the original csgA mutation. Group 2 contained two linked alleles that were unlinked to the csgA locus and restored fruiting-body formation, sporulation, and rippling. Group 3 revertants regained the ability to sporulate in fruiting bodies but not the ability to ripple. Revertants in groups 4 to 7 were able to sporulate but unable to form fruiting bodies or ripples. The suppressors were all found to be bypass suppressors even though they were not selected as such in most cases. The csgA mutation prevented expression of several developmentally regulated promoters, each fused to a lacZ reporter gene and assayed by beta-galactosidase production. In four of five suppressor groups (groups 4 to 7), expression of each of these csgA-dependent fusions was restored, which suggests that bypass suppression restores developmental gene expression near the point at which expression is disrupted in CsgA mutants. Bypass suppression did not restore production of C factor, and morphological manifestations of development such as rippling and fruiting-body formation were usually abnormal. One interpretation of these results is that C factor has multiple functions and few suppressors can compensate for all of them.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Atkinson K. D. SACCHAROMYCES CEREVISIAE Recessive Suppressor That Circumvents Phosphatidylserine Deficiency. Genetics. 1984 Nov;108(3):533–543. doi: 10.1093/genetics/108.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson K. D. Two recessive suppressors of Saccharomyces cerevisiae cho1 that are unlinked but fall in the same complementation group. Genetics. 1985 Sep;111(1):1–6. doi: 10.1093/genetics/111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson K., Fogel S., Henry S. A. Yeast mutant defective in phosphatidylserine synthesis. J Biol Chem. 1980 Jul 25;255(14):6653–6661. [PubMed] [Google Scholar]
- Avery L., Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191(1):99–109. doi: 10.1007/BF00330896. [DOI] [PubMed] [Google Scholar]
- Barbour S. D., Nagaishi H., Templin A., Clark A. J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. II. Rec+ revertants caused by indirect suppression of rec- mutations. Proc Natl Acad Sci U S A. 1970 Sep;67(1):128–135. doi: 10.1073/pnas.67.1.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Maurer R. Genetic approaches to the analysis of microbial development. Annu Rev Genet. 1982;16:61–83. doi: 10.1146/annurev.ge.16.120182.000425. [DOI] [PubMed] [Google Scholar]
- Clark A. J. The beginning of a genetic analysis of recombination proficiency. J Cell Physiol. 1967 Oct;70(2 Suppl):165–180. doi: 10.1002/jcp.1040700412. [DOI] [PubMed] [Google Scholar]
- DWORKIN M., GIBSON S. M. A SYSTEM FOR STUDYING MICROBIAL MORPHOGENESIS: RAPID FORMATION OF MICROCYSTS IN MYXOCOCCUS XANTHUS. Science. 1964 Oct 9;146(3641):243–244. doi: 10.1126/science.146.3641.243. [DOI] [PubMed] [Google Scholar]
- DWORKIN M. Nutritional requirements for vegetative growth of Myxococcus xanthus. J Bacteriol. 1962 Aug;84:250–257. doi: 10.1128/jb.84.2.250-257.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downard J. S., Zusman D. R. Differential expression of protein S genes during Myxococcus xanthus development. J Bacteriol. 1985 Mar;161(3):1146–1155. doi: 10.1128/jb.161.3.1146-1155.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M., Kaiser D. Cell interactions in myxobacterial growth and development. Science. 1985 Oct 4;230(4721):18–24. doi: 10.1126/science.3929384. [DOI] [PubMed] [Google Scholar]
- Englesberg E., Sheppard D., Squires C., Meronk F., Jr An analysis of "revertants" of a deletion mutant in the C gene of the L-arabinose gene complex in Escherichia coli B-r: isolation of initiator constitutive mutants (Ic). J Mol Biol. 1969 Jul 28;43(2):281–298. doi: 10.1016/0022-2836(69)90268-x. [DOI] [PubMed] [Google Scholar]
- Geisselsoder J., Campos J. M., Zusman D. R. Physical characterization of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):179–189. doi: 10.1016/0022-2836(78)90432-1. [DOI] [PubMed] [Google Scholar]
- Hagen D. C., Bretscher A. P., Kaiser D. Synergism between morphogenetic mutants of Myxococcus xanthus. Dev Biol. 1978 Jun;64(2):284–296. doi: 10.1016/0012-1606(78)90079-9. [DOI] [PubMed] [Google Scholar]
- Hodgkin J., Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye M., Inouye S., Zusman D. R. Biosynthesis and self-assembly of protein S, a development-specific protein of Myxococcus xanthus. Proc Natl Acad Sci U S A. 1979 Jan;76(1):209–213. doi: 10.1073/pnas.76.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser D. Control of multicellular development: Dictyostelium and Myxococcus. Annu Rev Genet. 1986;20:539–566. doi: 10.1146/annurev.ge.20.120186.002543. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Construction of Tn5 lac, a transposon that fuses lacZ expression to exogenous promoters, and its introduction into Myxococcus xanthus. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5816–5820. doi: 10.1073/pnas.81.18.5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroos L., Kaiser D. Expression of many developmentally regulated genes in Myxococcus depends on a sequence of cell interactions. Genes Dev. 1987 Oct;1(8):840–854. doi: 10.1101/gad.1.8.840. [DOI] [PubMed] [Google Scholar]
- Kroos L., Kuspa A., Kaiser D. A global analysis of developmentally regulated genes in Myxococcus xanthus. Dev Biol. 1986 Sep;117(1):252–266. doi: 10.1016/0012-1606(86)90368-4. [DOI] [PubMed] [Google Scholar]
- Kuner J. M., Kaiser D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc Natl Acad Sci U S A. 1981 Jan;78(1):425–429. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRossa R., Kuner J., Hagen D., Manoil C., Kaiser D. Developmental cell interactions of Myxococcus xanthus: analysis of mutants. J Bacteriol. 1983 Mar;153(3):1394–1404. doi: 10.1128/jb.153.3.1394-1404.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh N., Oosawa K., Simon M. I. Characterization of Escherichia coli chemotaxis receptor mutants with null phenotypes. J Bacteriol. 1986 Sep;167(3):992–998. doi: 10.1128/jb.167.3.992-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Asher S. J. Use of recombination techniques to examine the structure of the csg locus of Myxococcus xanthus. Mol Gen Genet. 1988 Jan;211(1):63–71. doi: 10.1007/BF00338394. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J. Control of morphogenesis in myxobacteria. Crit Rev Microbiol. 1987;14(3):195–227. doi: 10.3109/10408418709104439. [DOI] [PubMed] [Google Scholar]
- Shimkets L. J., Gill R. E., Kaiser D. Developmental cell interactions in Myxococcus xanthus and the spoC locus. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1406–1410. doi: 10.1073/pnas.80.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimkets L. J., Kaiser D. Induction of coordinated movement of Myxococcus xanthus cells. J Bacteriol. 1982 Oct;152(1):451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodergren E., Kaiser D. Insertions of Tn5 near genes that govern stimulatable cell motility in Myxococcus. J Mol Biol. 1983 Jun 25;167(2):295–310. doi: 10.1016/s0022-2836(83)80337-4. [DOI] [PubMed] [Google Scholar]
- Sudo S. Z., Dworkin M. Resistance of vegetative cells and microcysts of Myxococcus xanthus. J Bacteriol. 1969 Jun;98(3):883–887. doi: 10.1128/jb.98.3.883-887.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngman P., Zuber P., Perkins J. B., Sandman K., Igo M., Losick R. New ways to study developmental genes in spore-forming bacteria. Science. 1985 Apr 19;228(4697):285–291. doi: 10.1126/science.228.4697.285. [DOI] [PubMed] [Google Scholar]



