Abstract
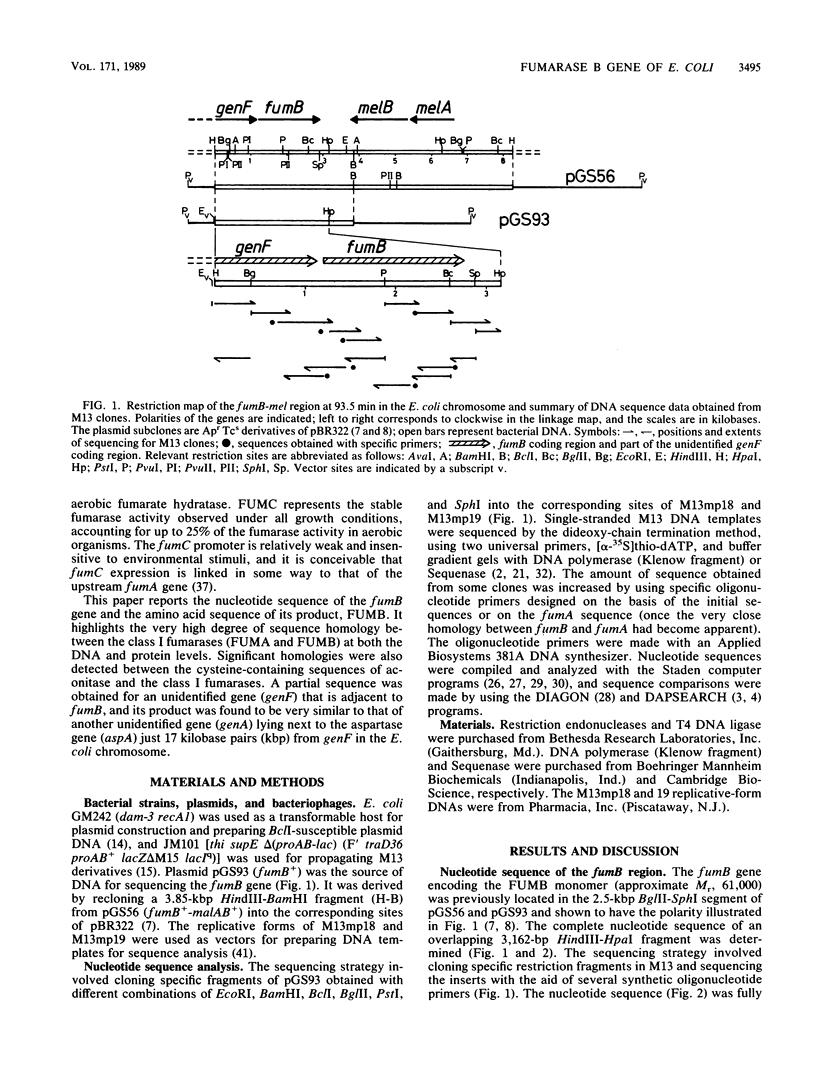
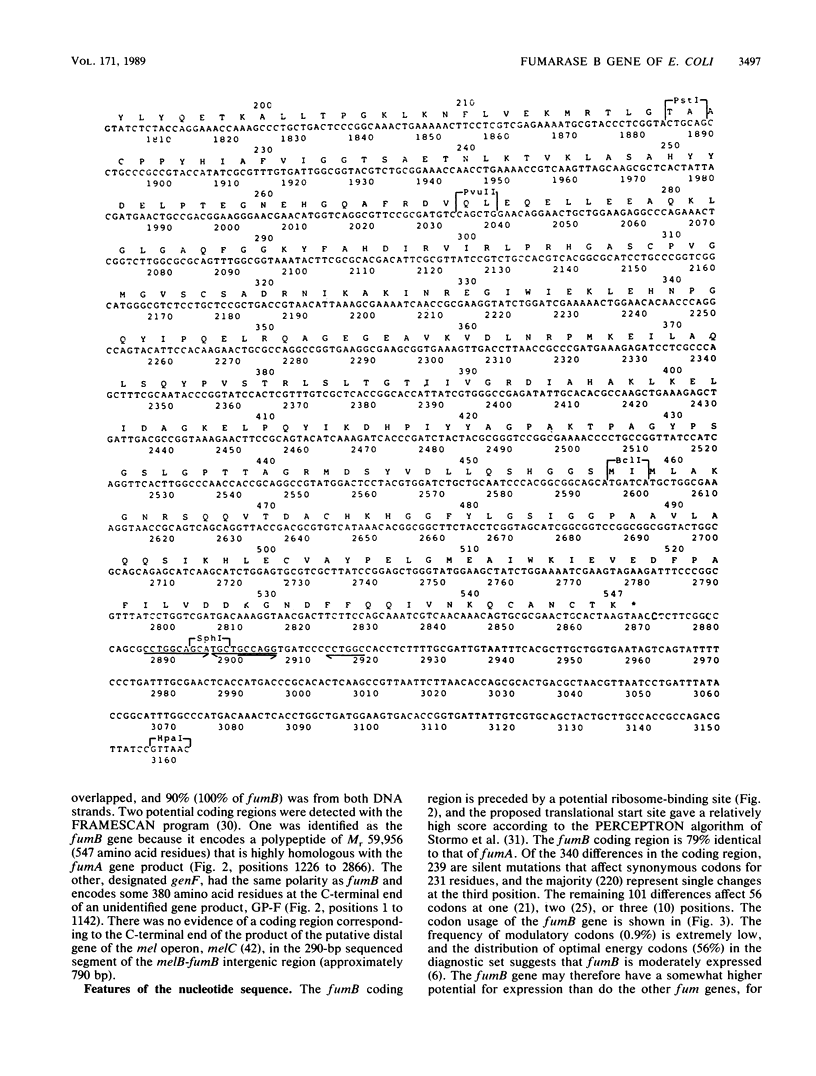
The nucleotide sequence of a 3,162-base-pair (bp) segment of DNA containing the FNR-regulated fumB gene, which encodes the anaerobic class I fumarase (FUMB) of Escherichia coli, was determined. The structural gene was found to comprise 1,641 bp, 547 codons (excluding the initiation and termination codons), and the gene product had a predicted Mr of 59,956. The amino acid sequence of FUMB contained the same number of residues as did that of the aerobic class I fumarase (FUMA), and there were identical amino acids at all but 56 positions (89.8% identity). There was no significant similarity between the class I fumarases and the class II enzyme (FUMC) except in one region containing the following consensus: Gly-Ser-Xxx-Ile-Met-Xxx-Xxx-Lys-Xxx-Asn. Some of the 56 amino acid substitutions must be responsible for the functional preferences of the enzymes for malate dehydration (FUMB) and fumarate hydration (FUMA). Significant similarities between the cysteine-containing sequence of the class I fumarases (FUMA and FUMB) and the mammalian aconitases were detected, and this finding further supports the view that these enzymes are all members of a family of iron-containing hydrolyases. The nucleotide sequence of a 1,142-bp distal sequence of an unidentified gene (genF) located upstream of fumB was also defined and found to encode a product that is homologous to the product of another unidentified gene (genA), located downstream of the neighboring aspartase gene (aspA).
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. F., Coulson A. F., Lyall A. The significance of protein sequence similarities. Comput Appl Biosci. 1988 Mar;4(1):67–71. doi: 10.1093/bioinformatics/4.1.67. [DOI] [PubMed] [Google Scholar]
- Darlison M. G., Guest J. R. Nucleotide sequence encoding the iron-sulphur protein subunit of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984 Oct 15;223(2):507–517. doi: 10.1042/bj2230507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Miles J. S., Roberts R. E., Woods S. A. The fumarase genes of Escherichia coli: location of the fumB gene and discovery of a new gene (fumC). J Gen Microbiol. 1985 Nov;131(11):2971–2984. doi: 10.1099/00221287-131-11-2971. [DOI] [PubMed] [Google Scholar]
- Guest J. R., Roberts R. E. Cloning, mapping, and expression of the fumarase gene of Escherichia coli K-12. J Bacteriol. 1983 Feb;153(2):588–596. doi: 10.1128/jb.153.2.588-596.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm K. S., Gawron O., Piszkiewicz D. Amino acid sequence of a peptide containing an essential cysteine residue of pig heart aconitase. Biochim Biophys Acta. 1981 Feb 27;667(2):457–461. doi: 10.1016/0005-2795(81)90211-7. [DOI] [PubMed] [Google Scholar]
- Henson J. M., Blake N. K., Marek L. E. The isolation of fumB mutants of Escherichia coli. J Gen Microbiol. 1987 Sep;133(9):2631–2638. doi: 10.1099/00221287-133-9-2631. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Marinus M. G., Morris N. R. Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12. J Bacteriol. 1973 Jun;114(3):1143–1150. doi: 10.1128/jb.114.3.1143-1150.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Complete nucleotide sequence of the fumarase gene fumA, of Escherichia coli. Nucleic Acids Res. 1984 Apr 25;12(8):3631–3642. doi: 10.1093/nar/12.8.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miles J. S., Guest J. R. Molecular genetic aspects of the citric acid cycle of Escherichia coli. Biochem Soc Symp. 1987;54:45–65. [PubMed] [Google Scholar]
- Plank D. W., Howard J. B. Identification of the reactive sulfhydryl and sequences of cysteinyl-tryptic peptides from beef heart aconitase. J Biol Chem. 1988 Jun 15;263(17):8184–8189. [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sacchettini J. C., Frazier M. W., Chiara D. C., Banaszak L. J., Grant G. A. Amino acid sequence of porcine heart fumarase. Biochem Biophys Res Commun. 1988 May 31;153(1):435–440. doi: 10.1016/s0006-291x(88)81243-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Schweiger G., Dutscho R., Buckel W. Purification of 2-hydroxyglutaryl-CoA dehydratase from Acidaminococcus fermentans. An iron-sulfur protein. Eur J Biochem. 1987 Dec 1;169(2):441–448. doi: 10.1111/j.1432-1033.1987.tb13631.x. [DOI] [PubMed] [Google Scholar]
- Shaw D. J., Rice D. W., Guest J. R. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol. 1983 May 15;166(2):241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. Activation of the lac operon of Escherichia coli by a mutant FNR protein. Mol Microbiol. 1987 Jul;1(1):53–58. doi: 10.1111/j.1365-2958.1987.tb00526.x. [DOI] [PubMed] [Google Scholar]
- Spiro S., Guest J. R. Regulation and over-expression of the fnr gene of Escherichia coli. J Gen Microbiol. 1987 Dec;133(12):3279–3288. doi: 10.1099/00221287-133-12-3279. [DOI] [PubMed] [Google Scholar]
- Staden R. A new computer method for the storage and manipulation of DNA gel reading data. Nucleic Acids Res. 1980 Aug 25;8(16):3673–3694. doi: 10.1093/nar/8.16.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. A strategy of DNA sequencing employing computer programs. Nucleic Acids Res. 1979 Jun 11;6(7):2601–2610. doi: 10.1093/nar/6.7.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. An interactive graphics program for comparing and aligning nucleic acid and amino acid sequences. Nucleic Acids Res. 1982 May 11;10(9):2951–2961. doi: 10.1093/nar/10.9.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):505–519. doi: 10.1093/nar/12.1part2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J. S., Ida N., Tokushige M., Sakamoto H., Shimura Y. Cloning and nucleotide sequence of the aspartase gene of Escherichia coli W. Nucleic Acids Res. 1985 Mar 25;13(6):2063–2074. doi: 10.1093/nar/13.6.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wilde R. J., Guest J. R. Transcript analysis of the citrate synthase and succinate dehydrogenase genes of Escherichia coli K12. J Gen Microbiol. 1986 Dec;132(12):3239–3251. doi: 10.1099/00221287-132-12-3239. [DOI] [PubMed] [Google Scholar]
- Wood D., Darlison M. G., Wilde R. J., Guest J. R. Nucleotide sequence encoding the flavoprotein and hydrophobic subunits of the succinate dehydrogenase of Escherichia coli. Biochem J. 1984 Sep 1;222(2):519–534. doi: 10.1042/bj2220519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods S. A., Miles J. S., Roberts R. E., Guest J. R. Structural and functional relationships between fumarase and aspartase. Nucleotide sequences of the fumarase (fumC) and aspartase (aspA) genes of Escherichia coli K12. Biochem J. 1986 Jul 15;237(2):547–557. doi: 10.1042/bj2370547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods S. A., Schwartzbach S. D., Guest J. R. Two biochemically distinct classes of fumarase in Escherichia coli. Biochim Biophys Acta. 1988 Apr 28;954(1):14–26. doi: 10.1016/0167-4838(88)90050-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yazyu H., Shiota-Niiya S., Shimamoto T., Kanazawa H., Futai M., Tsuchiya T. Nucleotide sequence of the melB gene and characteristics of deduced amino acid sequence of the melibiose carrier in Escherichia coli. J Biol Chem. 1984 Apr 10;259(7):4320–4326. [PubMed] [Google Scholar]
- Yumoto N., Tokushige M. Characterization of multiple fumarase proteins in Escherichia coli. Biochem Biophys Res Commun. 1988 Jun 30;153(3):1236–1243. doi: 10.1016/s0006-291x(88)81360-3. [DOI] [PubMed] [Google Scholar]


