Abstract
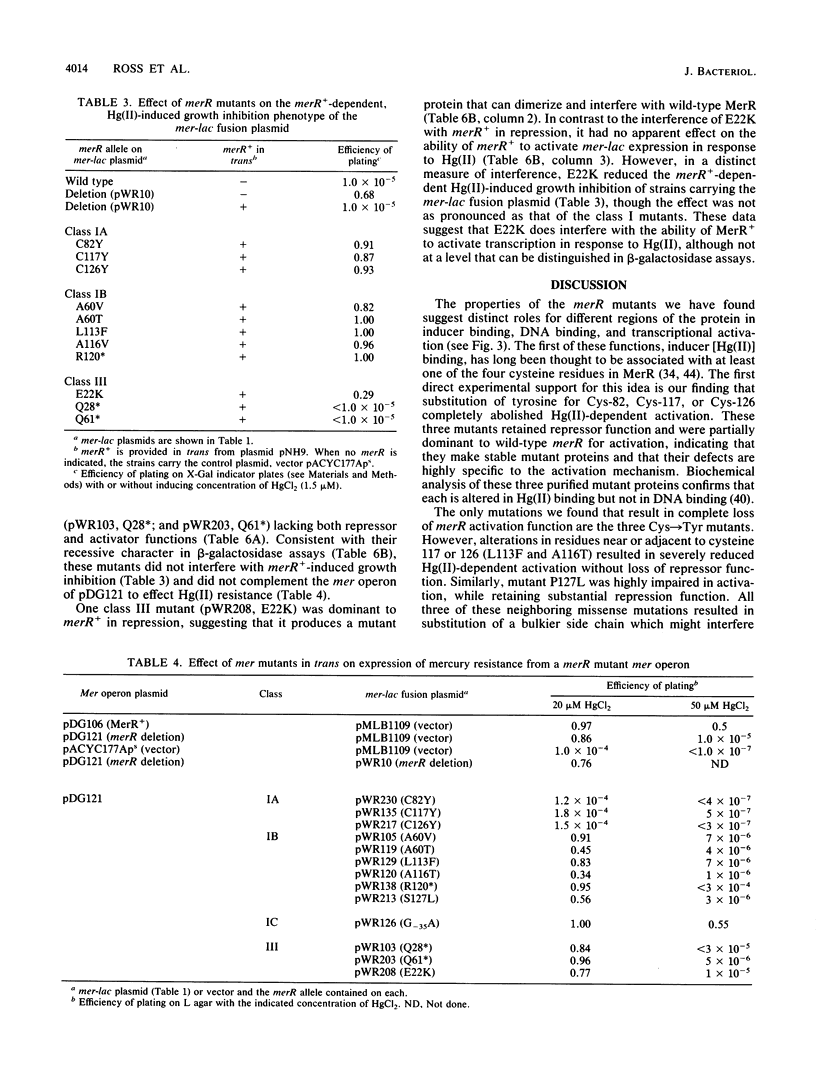
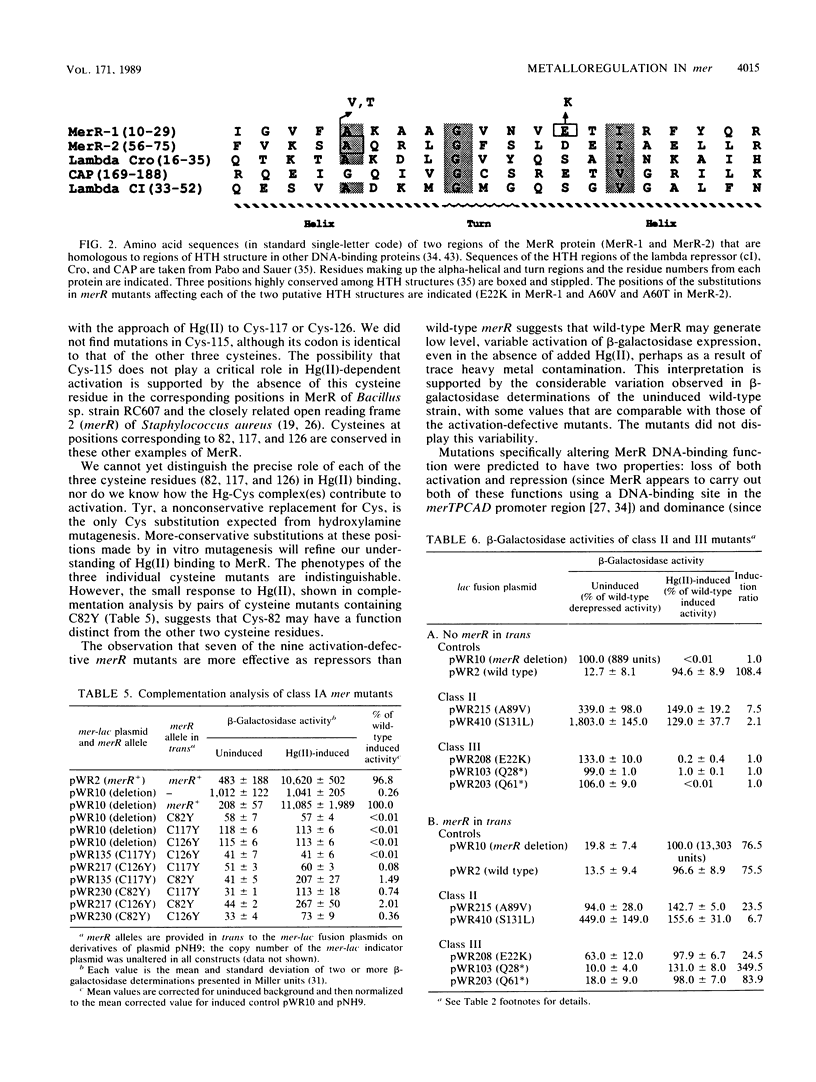
Transcription of the Tn21 mercury resistance operon (mer) is controlled by the toxic metal cation Hg(II). This control is mediated by the product of the merR gene, a 144-amino-acid protein which represses transcription of the structural genes (merTPCAD) in the absence of Hg(II) and activates transcription in the presence of Hg(II). We have used a mer-lac transcriptional fusion to obtain regulatory mutants in this metal-responsive system. Some mutants were defective in Hg(II)-induced activation while retaining repression function (a- r+), others were defective in repression but not activation (a+ r-), and some had lost both functions (a- r-). Mutations in three of the four cysteine residues of merR resulted in complete loss of Hg(II)-inducible activation but retention of the repressor function, suggesting that these residues serve as ligands for Hg(II) in the activation process. Other lesions adjacent to or very near these cysteines exhibited severely reduced activation and also retained repressor function. There were two putative helix-turn-helix (HTH) domains in merR, and mutants in each had very different phenotypes. A partially dominant mutation in the more amino-terminal region of the two putative HTH regions resulted in loss of both activation and repression (a- r-), consistent with a role for this region in DNA binding. Mutations in the more centrally located HTH region resulted only in loss of Hg(II)-induced activation (a- r+). Lesions in the central and in the carboxy-terminal regions of merR exhibited both Hg(II)-independent and Hg(II)-dependent transcriptional activation, suggesting that elements important in the activation mechanism may be widely distributed in this relatively small protein. The sole cis-acting mutant obtained with this operon fusion strategy, a down-promoter mutation, lies in a highly conserved base in the -35 region of the merTPCAD promoter.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Nakamura T., Mitani H., Mori H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3329–3332. doi: 10.1002/j.1460-2075.1985.tb04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrineau P., Gilbert P., Jackson W. J., Jones C. S., Summers A. O., Wisdom S. The DNA sequence of the mercury resistance operon of the IncFII plasmid NR1. J Mol Appl Genet. 1984;2(6):601–619. [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Ptashne M. Activation of transcription by the bacteriophage 434 repressor. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9353–9357. doi: 10.1073/pnas.83.24.9353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman F. D., Ptashne M. Turning lambda Cro into a transcriptional activator. Cell. 1988 Jul 15;54(2):191–197. doi: 10.1016/0092-8674(88)90551-x. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Brown N. L. Identification of the merR gene of R100 by using mer-lac gene and operon fusions. J Bacteriol. 1985 Sep;163(3):1153–1157. doi: 10.1128/jb.163.3.1153-1157.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Ginnity F. Some mercurial resistance plasmids from different incompatibility groups specify merR regulatory functions that both repress and induce the mer operon of plasmid R100. J Bacteriol. 1985 May;162(2):773–776. doi: 10.1128/jb.162.2.773-776.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H., Weiss A. A., Silver S. Transposon A-generated mutations in the mercuric resistance genes of plasmid R100-1. J Bacteriol. 1979 Oct;140(1):167–181. doi: 10.1128/jb.140.1.167-181.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. The genetics and biochemistry of mercury resistance. Crit Rev Microbiol. 1987;15(2):117–140. doi: 10.3109/10408418709104455. [DOI] [PubMed] [Google Scholar]
- Gambill B. D., Summers A. O. Versatile mercury-resistant cloning and expression vectors. Gene. 1985;39(2-3):293–297. doi: 10.1016/0378-1119(85)90326-9. [DOI] [PubMed] [Google Scholar]
- Garges S., Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985 Jul;41(3):745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Griffin H. G., Foster T. J., Silver S., Misra T. K. Cloning and DNA sequence of the mercuric- and organomercurial-resistance determinants of plasmid pDU1358. Proc Natl Acad Sci U S A. 1987 May;84(10):3112–3116. doi: 10.1073/pnas.84.10.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht M. H., Sauer R. T. Phage lambda repressor revertants. Amino acid substitutions that restore activity to mutant proteins. J Mol Biol. 1985 Nov 5;186(1):53–63. doi: 10.1016/0022-2836(85)90256-6. [DOI] [PubMed] [Google Scholar]
- Helmann J. D., Wang Y., Mahler I., Walsh C. T. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. 1989 Jan;171(1):222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltzel A., Gambill D., Jackson W. J., Totis P. A., Summers A. O. Overexpression and DNA-binding properties of the mer-encoded regulatory protein from plasmid NR1 (Tn21). J Bacteriol. 1987 Jul;169(7):3379–3384. doi: 10.1128/jb.169.7.3379-3384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Hochschild A., Ptashne M. Homologous interactions of lambda repressor and lambda Cro with the lambda operator. Cell. 1986 Mar 28;44(6):925–933. doi: 10.1016/0092-8674(86)90015-2. [DOI] [PubMed] [Google Scholar]
- Irwin N., Ptashne M. Mutants of the catabolite activator protein of Escherichia coli that are specifically deficient in the gene-activation function. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8315–8319. doi: 10.1073/pnas.84.23.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isackson P. J., Bertrand K. P. Dominant negative mutations in the Tn10 tet repressor: evidence for use of the conserved helix-turn-helix motif in DNA binding. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6226–6230. doi: 10.1073/pnas.82.18.6226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley R. L., Yanofsky C. Mutational studies with the trp repressor of Escherichia coli support the helix-turn-helix model of repressor recognition of operator DNA. Proc Natl Acad Sci U S A. 1985 Jan;82(2):483–487. doi: 10.1073/pnas.82.2.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laddaga R. A., Chu L., Misra T. K., Silver S. Nucleotide sequence and expression of the mercurial-resistance operon from Staphylococcus aureus plasmid pI258. Proc Natl Acad Sci U S A. 1987 Aug;84(15):5106–5110. doi: 10.1073/pnas.84.15.5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. A., Brown N. L. Regulation of transcription in Escherichia coli from the mer and merR promoters in the transposon Tn501. J Mol Biol. 1989 Jan 20;205(2):343–353. doi: 10.1016/0022-2836(89)90345-8. [DOI] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nelson H. C., Sauer R. T. Interaction of mutant lambda repressors with operator and non-operator DNA. J Mol Biol. 1986 Nov 5;192(1):27–38. doi: 10.1016/0022-2836(86)90461-4. [DOI] [PubMed] [Google Scholar]
- Ni'Bhriain N. N., Silver S., Foster T. J. Tn5 insertion mutations in the mercuric ion resistance genes derived from plasmid R100. J Bacteriol. 1983 Aug;155(2):690–703. doi: 10.1128/jb.155.2.690-703.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Halloran T., Walsh C. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science. 1987 Jan 9;235(4785):211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Ross W., Shore S. H., Howe M. M. Mutants of Escherichia coli defective for replicative transposition of bacteriophage Mu. J Bacteriol. 1986 Sep;167(3):905–919. doi: 10.1128/jb.167.3.905-919.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K., Beck C. F. Promoter-probe vectors for the analysis of divergently arranged promoters. Gene. 1986;42(1):37–48. doi: 10.1016/0378-1119(86)90148-4. [DOI] [PubMed] [Google Scholar]
- Shewchuk L. M., Helmann J. D., Ross W., Park S. J., Summers A. O., Walsh C. T. Transcriptional switching by the MerR protein: activation and repression mutants implicate distinct DNA and mercury(II) binding domains. Biochemistry. 1989 Mar 7;28(5):2340–2344. doi: 10.1021/bi00431a053. [DOI] [PubMed] [Google Scholar]
- Shewchuk L. M., Verdine G. L., Walsh C. T. Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury (II) binding activities. Biochemistry. 1989 Mar 7;28(5):2331–2339. doi: 10.1021/bi00431a052. [DOI] [PubMed] [Google Scholar]
- Simons R. W., Houman F., Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53(1):85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Summers A. O. Organization, expression, and evolution of genes for mercury resistance. Annu Rev Microbiol. 1986;40:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- Walsh C. T., Distefano M. D., Moore M. J., Shewchuk L. M., Verdine G. L. Molecular basis of bacterial resistance to organomercurial and inorganic mercuric salts. FASEB J. 1988 Feb;2(2):124–130. doi: 10.1096/fasebj.2.2.3277886. [DOI] [PubMed] [Google Scholar]
- Wang Y., Moore M., Levinson H. S., Silver S., Walsh C., Mahler I. Nucleotide sequence of a chromosomal mercury resistance determinant from a Bacillus sp. with broad-spectrum mercury resistance. J Bacteriol. 1989 Jan;171(1):83–92. doi: 10.1128/jb.171.1.83-92.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


