Abstract
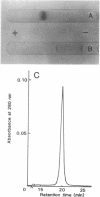
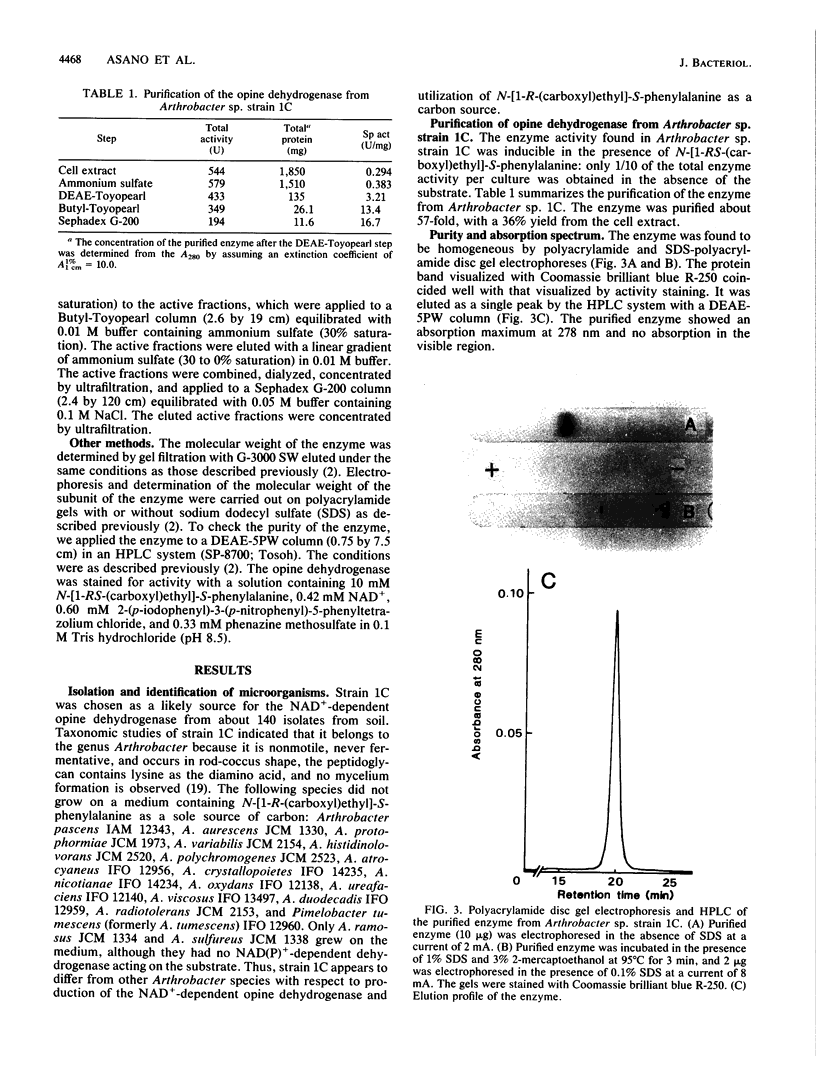
A new NAD+-dependent opine dehydrogenase was purified to homogeneity from Arthrobacter sp. strain 1C isolated from soil by an enrichment culture technique. The enzyme has a molecular weight of about 70,000 and consists of two identical subunits with molecular weights of about 36,000. The enzyme catalyzed a reversible oxidation-reduction reaction of opine-type secondary amine dicarboxylic acids. In the oxidative deamination reaction, the enzyme was active toward unusual opines, such as N-[1-R-(carboxyl)ethyl]-S-methionine and N-[1-R-(carboxyl)ethyl]-S-phenylalanine. In the reductive secondary amine-forming reaction with NADH as a cofactor, the enzyme utilized L-amino acids such as L-methionine, L-isoleucine, L-valine, L-phenylalanine, L-leucine, L-alanine, and L-threonine as amino donors and alpha-keto acids such as pyruvate, oxaloacetate, glyoxylate, and alpha-ketobutyrate as amino acceptors. The product enzymatically synthesized from L-phenylalanine and pyruvate in the presence of NADH was identified as N-[1-R-(carboxyl)ethyl]-S-phenylalanine.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asano Y., Nakazawa A., Endo K., Hibino Y., Ohmori M., Numao N., Kondo K. Phenylalanine dehydrogenase of Bacillus badius. Purification, characterization and gene cloning. Eur J Biochem. 1987 Oct 1;168(1):153–159. doi: 10.1111/j.1432-1033.1987.tb13399.x. [DOI] [PubMed] [Google Scholar]
- Asano Y., Nakazawa A., Endo K. Novel phenylalanine dehydrogenases from Sporosarcina ureae and Bacillus sphaericus. Purification and characterization. J Biol Chem. 1987 Jul 25;262(21):10346–10354. [PubMed] [Google Scholar]
- Asano Y., Sekigawa T., Inukai H., Nakazawa A. Purification and properties of formate dehydrogenase from Moraxella sp. strain C-1. J Bacteriol. 1988 Jul;170(7):3189–3193. doi: 10.1128/jb.170.7.3189-3193.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates H. A., Kaushal A., Deng P. N., Sciaky D. Structure and synthesis of histopine, a histidine derivative produced by crown gall tumors. Biochemistry. 1984 Jul 3;23(14):3287–3290. doi: 10.1021/bi00309a026. [DOI] [PubMed] [Google Scholar]
- Chang C. C., Chen C. M., Adams B. R., Trost B. M. Leucinopine, a characteristic compound of some crown-gall tumors. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3573–3576. doi: 10.1073/pnas.80.12.3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Greve H., Dhaese P., Seurinck J., Lemmers M., Van Montagu M., Schell J. Nucleotide sequence and transcript map of the Agrobacterium tumefaciens Ti plasmid-encoded octopine synthase gene. J Mol Appl Genet. 1982;1(6):499–511. [PubMed] [Google Scholar]
- Fields J. H., Eng A. K., Ramsden W. D., Hochachka P. W., Weinstein B. Alanopine and strombine are novel imino acids produced by a dehydrogenase found in the adductor muscle of the oyster, Crassostrea gigas. Arch Biochem Biophys. 1980 Apr 15;201(1):110–114. doi: 10.1016/0003-9861(80)90493-2. [DOI] [PubMed] [Google Scholar]
- Firmin J. L., Stewart I. M., Wilson K. E. N2-(1-carboxyethyl)methionine. A 'pseudo-opine' in octopine-type crown-gall tumours. Biochem J. 1985 Dec 1;232(2):431–434. doi: 10.1042/bj2320431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hack E., Kemp J. D. Comparison of octopine, histopine, lysopine, and octopinic acid synthesizing activities in sunflower crown gall tissues. Biochem Biophys Res Commun. 1977 Sep 23;78(2):785–791. doi: 10.1016/0006-291x(77)90248-0. [DOI] [PubMed] [Google Scholar]
- Hack E., Kemp J. D. Purification and Characterization of the Crown Gall-specific Enzyme, Octopine Synthase. Plant Physiol. 1980 May;65(5):949–955. doi: 10.1104/pp.65.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jubier M. -F. Degradation of lysopine by an inducible membrane-bound oxidase in Agrobacterium tumefaciens. FEBS Lett. 1972 Dec 1;28(2):129–132. doi: 10.1016/0014-5793(72)80693-8. [DOI] [PubMed] [Google Scholar]
- Kemp J. D., Sutton D. W., Hack E. Purification and characterization of the crown gall specific enzyme nopaline synthase. Biochemistry. 1979 Aug 21;18(17):3755–3760. doi: 10.1021/bi00584a017. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Misono H., Soda K. Properties of meso-alpha,epsilon-diaminopimelate D-dehydrogenase from Bacillus sphaericus. J Biol Chem. 1980 Nov 25;255(22):10599–10605. [PubMed] [Google Scholar]
- Montoya A. L., Chilton M. D., Gordon M. P., Sciaky D., Nester E. W. Octopine and nopaline metabolism in Agrobacterium tumefaciens and crown gall tumor cells: role of plasmid genes. J Bacteriol. 1977 Jan;129(1):101–107. doi: 10.1128/jb.129.1.101-107.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki N., Hibino Y., Asano Y., Ohmori M., Numao N., Kondo K. Cloning and nucleotide sequencing of phenylalanine dehydrogenase gene of Bacillus sphaericus. Gene. 1988 Mar 31;63(2):337–341. doi: 10.1016/0378-1119(88)90537-9. [DOI] [PubMed] [Google Scholar]
- Otten L. A., Vreugdenhil D., Schilperoort R. A. Properties of D(+)-lysopine dehydrogenase from crown gall tumour tissue. Biochim Biophys Acta. 1977 Dec 8;485(2):268–277. doi: 10.1016/0005-2744(77)90163-2. [DOI] [PubMed] [Google Scholar]
- Schardl C. L., Kado C. I. A functional map of the nopaline catabolism genes on the Ti plasmid of Agrobacterium tumefaciens C58. Mol Gen Genet. 1983;191(1):10–16. doi: 10.1007/BF00330882. [DOI] [PubMed] [Google Scholar]
- Schrimsher J. L., Taylor K. B. Octopine dehydrogenase from crown gall tumor and from Pecten maximus. Oxidation of (4R)- and (4S)-[4-3H]NADH. J Biol Chem. 1982 Aug 10;257(15):8953–8956. [PubMed] [Google Scholar]
- Storey K. B., Storey J. M. Kinetic characterization of tissue-specific isozymes of octopine dehydrogenase from mantle muscle and brain of Sepia officinalis. Functional similarities to the M4 and H4 isozymes of lactate dehydrogenase. Eur J Biochem. 1979 Feb 1;93(3):545–542. doi: 10.1111/j.1432-1033.1979.tb12853.x. [DOI] [PubMed] [Google Scholar]
- Tremblay G., Gagliardo R., Chilton W. S., Dion P. Diversity among Opine-Utilizing Bacteria: Identification of Coryneform Isolates. Appl Environ Microbiol. 1987 Jul;53(7):1519–1524. doi: 10.1128/aem.53.7.1519-1524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettlmeissl G., Teschner W., Rudolph R., Jaenicke R., Gäde G. Isolation, physicochemical properties, and folding of octopine dehydrogenase from Pecten jacobaeus. Eur J Biochem. 1984 Sep 3;143(2):401–407. doi: 10.1111/j.1432-1033.1984.tb08387.x. [DOI] [PubMed] [Google Scholar]
- van Thoai N., Huc C., Pho D. B., Olomucki A. Octopine déshydrogénase. Purification et propriétés catalytiques. Biochim Biophys Acta. 1969 Sep 30;191(1):46–57. doi: 10.1016/0005-2744(69)90313-1. [DOI] [PubMed] [Google Scholar]