Abstract
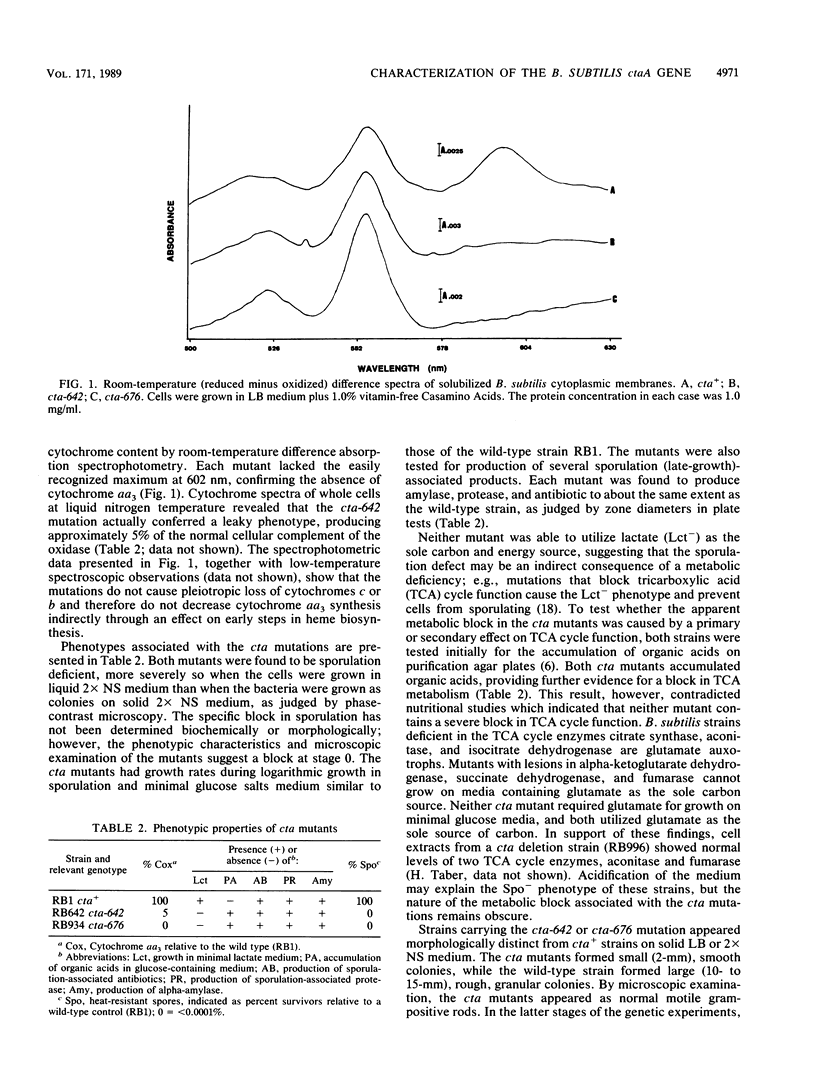
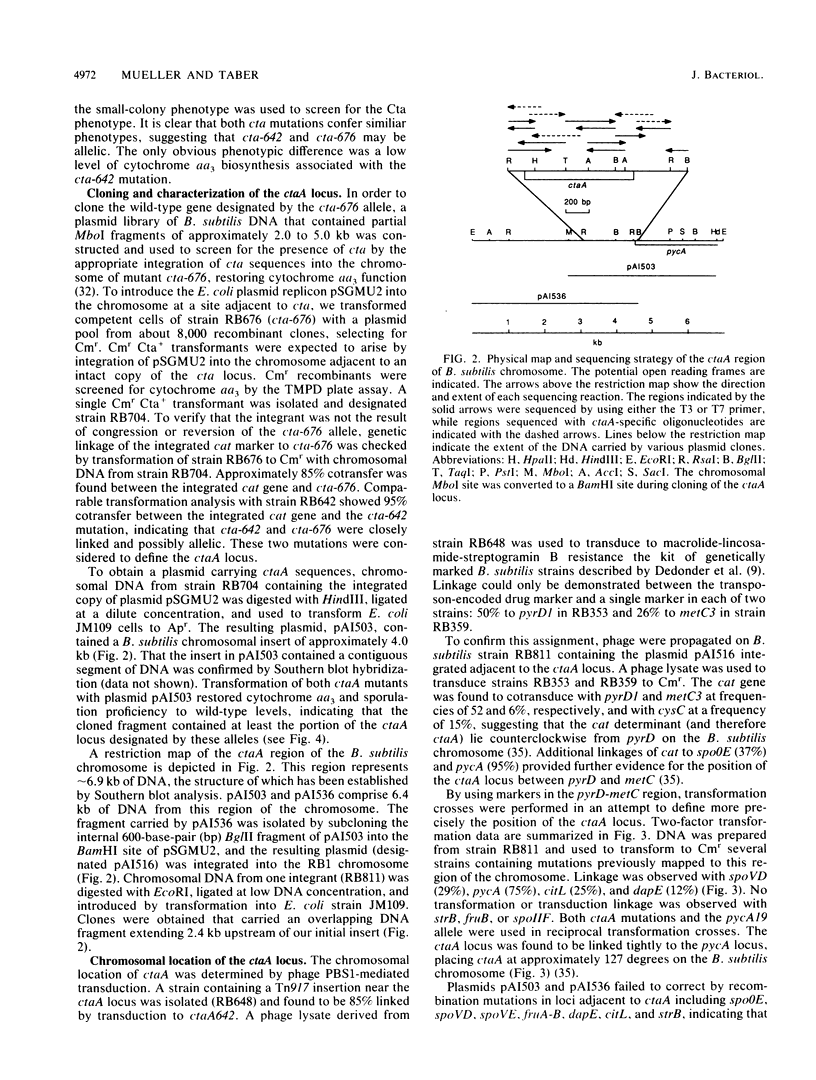
Cytochrome aa3 is one of two terminal oxidase complexes in the Bacillus subtilis electron transport chain. A novel genetic strategy was devised which permitted the isolation of B. subtilis mutants lacking cytochrome aa3 by selection for streptomycin-resistant clones which failed to oxidize the artificial electron donor N,N,N',N'-tetramethyl-p-phenylenediamine. Two mutations were studied intensively. Spectroscopic examination showed that each mutant lacked cytochrome aa3; they were also asporogenous and unable to grow on lactate as the sole carbon and energy source. These mutations were mapped to a locus designated ctaA, located at 127 degrees between pyrD and metC on the B. subtilis chromosome. Both ctaA mutations were closely linked by transformation to the pycA locus. The ctaA locus and a portion of the pycA locus were cloned from a B. subtilis integration library constructed in Escherichia coli. A recombinant plasmid containing a 4.0-kilobase insert of B. subtilis DNA could transform both ctaA mutants to CtaA+. Gene disruption and complementation experiments with subcloned fragments revealed that the ctaA locus consisted of a single transcriptional unit about 1.35 kilobase pairs in size. The nucleotide sequence of the ctaA transcriptional unit contains a single open reading frame capable of coding for a protein with a predicted molecular weight of 34,065. The predicted protein is extremely hydrophobic, with several probable membrane-spanning domains. No sequence similiarity was found between ctaA and the highly conserved procaryotic and mitochondrial oxidase polypeptides. Cloning and sequence analysis of two ctaA mutations revealed that one allele is a nonsense mutation in the carboxy terminus and the other is a missense mutation in the amino terminus; this indicates that the pleiotropic phenotype conferred by each mutation was caused by loss of CtaA or of its activity. Genetic evidence suggests that the ctaA gene product is required as an accessory protein in the genetic expression, posttranslational biogenesis, or both, of the cytochrome aa3 complex and during an early stage of sporogenesis.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrow A. S., Taber H. W. Streptomycin accumulation by Bacillus subtilis requires both a membrane potential and cytochrome aa3. Antimicrob Agents Chemother. 1986 Jan;29(1):141–146. doi: 10.1128/aac.29.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisschop A., Konings W. N. Reconstitution of reduced nicotinamide adenine dinucleotide oxidase activity with menadione in membrane vesicles from the menaquinone-deficient Bacillus subtilis aro D. Relation between electron transfer and active transport. Eur J Biochem. 1976 Aug 16;67(2):357–365. doi: 10.1111/j.1432-1033.1976.tb10699.x. [DOI] [PubMed] [Google Scholar]
- Buxton R. S. A heat-sensitive lysis mutant of Bacillus subtilis 168 with a low activity of pyruvate carboxylase. J Gen Microbiol. 1978 Apr;105(2):175–185. doi: 10.1099/00221287-105-2-175. [DOI] [PubMed] [Google Scholar]
- Carls R. A., Hanson R. S. Isolation and characterization of tricarboxylic acid cycle mutants of Bacillus subtilis. J Bacteriol. 1971 Jun;106(3):848–855. doi: 10.1128/jb.106.3.848-855.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Contente S., Dubnau D. Characterization of plasmid transformation in Bacillus subtilis: kinetic properties and the effect of DNA conformation. Mol Gen Genet. 1979 Jan 2;167(3):251–258. doi: 10.1007/BF00267416. [DOI] [PubMed] [Google Scholar]
- De Vrij W., Azzi A., Konings W. N. Structural and functional properties of cytochrome c oxidase from Bacillus subtilis W23. Eur J Biochem. 1983 Mar 1;131(1):97–103. doi: 10.1111/j.1432-1033.1983.tb07235.x. [DOI] [PubMed] [Google Scholar]
- Dedonder R. A., Lepesant J. A., Lepesant-Kejzlarová J., Billault A., Steinmetz M., Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol. 1977 Apr;33(4):989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diesterhaft M. D., Freese E. Role of pyruvate carboxylase, phosphoenolpyruvate carboxykinase, and malic enzyme during growth and sporulation of Bacillus subtilis. J Biol Chem. 1973 Sep 10;248(17):6062–6070. [PubMed] [Google Scholar]
- Edwards C., Beer S., Siviram A., Chance B. Photochemical action spectra of bacterial a- and o-type oxidases using a dye laser. FEBS Lett. 1981 Jun 15;128(2):205–207. doi: 10.1016/0014-5793(81)80081-6. [DOI] [PubMed] [Google Scholar]
- Errington J. Efficient Bacillus subtilis cloning system using bacteriophage vector phi 105J9. J Gen Microbiol. 1984 Oct;130(10):2615–2628. doi: 10.1099/00221287-130-10-2615. [DOI] [PubMed] [Google Scholar]
- Fort P., Errington J. Nucleotide sequence and complementation analysis of a polycistronic sporulation operon, spoVA, in Bacillus subtilis. J Gen Microbiol. 1985 May;131(5):1091–1105. doi: 10.1099/00221287-131-5-1091. [DOI] [PubMed] [Google Scholar]
- Fortnagel P., Freese E. Analysis of sporulation mutants. II. Mutants blocked in the citric acid cycle. J Bacteriol. 1968 Apr;95(4):1431–1438. doi: 10.1128/jb.95.4.1431-1438.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green G. N., Gennis R. B. Isolation and characterization of an Escherichia coli mutant lacking cytochrome d terminal oxidase. J Bacteriol. 1983 Jun;154(3):1269–1275. doi: 10.1128/jb.154.3.1269-1275.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D., Errington J. Construction of improved bacteriophage phi 105 vectors for cloning by transfection in Bacillus subtilis. J Gen Microbiol. 1987 Mar;133(3):483–492. doi: 10.1099/00221287-133-3-483. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Leighton T. J., Doi R. H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971 May 25;246(10):3189–3195. [PubMed] [Google Scholar]
- McEnroe A. S., Taber H. W. Correlation between cytochrome aa3 concentrations and streptomycin accumulation in Bacillus subtilis. Antimicrob Agents Chemother. 1984 Oct;26(4):507–512. doi: 10.1128/aac.26.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierendorf R. C., Pfeffer D. Direct sequencing of denatured plasmid DNA. Methods Enzymol. 1987;152:556–562. doi: 10.1016/0076-6879(87)52061-4. [DOI] [PubMed] [Google Scholar]
- Miller P., Mueller J., Hill K., Taber H. Transcriptional regulation of a promoter in the men gene cluster of Bacillus subtilis. J Bacteriol. 1988 Jun;170(6):2742–2748. doi: 10.1128/jb.170.6.2742-2748.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niaudet B., Goze A., Ehrlich S. D. Insertional mutagenesis in Bacillus subtilis: mechanism and use in gene cloning. Gene. 1982 Oct;19(3):277–284. doi: 10.1016/0378-1119(82)90017-8. [DOI] [PubMed] [Google Scholar]
- O'brian M. R., Maier R. J. Isolation of a cytochrome aa(3) gene from Bradyrhizobium japonicum. Proc Natl Acad Sci U S A. 1987 May;84(10):3219–3223. doi: 10.1073/pnas.84.10.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Curtis C. A., de Lencastre H. Use of integrational plasmid vectors to demonstrate the polycistronic nature of a transcriptional unit (spoIIA) required for sporulation of Bacillus subtilis. J Gen Microbiol. 1984 Aug;130(8):2123–2136. doi: 10.1099/00221287-130-8-2123. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole R. K. Bacterial cytochrome oxidases. A structurally and functionally diverse group of electron-transfer proteins. Biochim Biophys Acta. 1983 Sep 15;726(3):205–243. doi: 10.1016/0304-4173(83)90006-x. [DOI] [PubMed] [Google Scholar]
- Raitio M., Jalli T., Saraste M. Isolation and analysis of the genes for cytochrome c oxidase in Paracoccus denitrificans. EMBO J. 1987 Sep;6(9):2825–2833. doi: 10.1002/j.1460-2075.1987.tb02579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C. W., Schmidt B. J., Mirot M. S., Thompson L. D., Guyer M. S. Use of chromosomal integration in the establishment and expression of blaZ, a Staphylococcus aureus beta-lactamase gene, in Bacillus subtilis. J Bacteriol. 1984 Mar;157(3):718–726. doi: 10.1128/jb.157.3.718-726.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short J. M., Fernandez J. M., Sorge J. A., Huse W. D. Lambda ZAP: a bacteriophage lambda expression vector with in vivo excision properties. Nucleic Acids Res. 1988 Aug 11;16(15):7583–7600. doi: 10.1093/nar/16.15.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone N., Yokoi F., Fu T., Ohta S., Metso T., Raitio M., Saraste M. Nucleotide sequence of the gene coding for cytochrome oxidase subunit I from the thermophilic bacterium PS3. J Biochem. 1988 Apr;103(4):606–610. doi: 10.1093/oxfordjournals.jbchem.a122314. [DOI] [PubMed] [Google Scholar]
- Staal S. P., Hoch J. A. Conditional dihydrostreptomycin resistance in Bacillus subtilis. J Bacteriol. 1972 Apr;110(1):202–207. doi: 10.1128/jb.110.1.202-207.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinrücke P., Steffens G. C., Panskus G., Buse G., Ludwig B. Subunit II of cytochrome c oxidase from Paracoccus denitrificans. DNA sequence, gene expression and the protein. Eur J Biochem. 1987 Sep 15;167(3):431–439. doi: 10.1111/j.1432-1033.1987.tb13356.x. [DOI] [PubMed] [Google Scholar]
- Sullivan M. A., Yasbin R. E., Young F. E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984 Jul-Aug;29(1-2):21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- Taber H. W., Dellers E. A., Lombardo L. R. Menaquinone biosynthesis in Bacillus subtilis: isolation of men mutants and evidence for clustering of men genes. J Bacteriol. 1981 Jan;145(1):321–327. doi: 10.1128/jb.145.1.321-327.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. W., Mueller J. P., Miller P. F., Arrow A. S. Bacterial uptake of aminoglycoside antibiotics. Microbiol Rev. 1987 Dec;51(4):439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. Isolation and properties of cytochrome a deficient mutants of Bacillus subtilis. J Gen Microbiol. 1974 Apr;81(2):435–444. doi: 10.1099/00221287-81-2-435. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Trowsdale J., Chen S. M., Hoch J. A. Genetic analysis of a class of polymyxin resistant partial revertants of stage O sporulation mutants of Bacillus subtilis: map of the chromosome region near the origin of replication. Mol Gen Genet. 1979 May 23;173(1):61–70. doi: 10.1007/BF00267691. [DOI] [PubMed] [Google Scholar]
- de Vrij W., Driessen A. J., Hellingwerf K. J., Konings W. N. Measurements of the proton motive force generated by cytochrome c oxidase from Bacillus subtilis in proteoliposomes and membrane vesicles. Eur J Biochem. 1986 Apr 15;156(2):431–440. doi: 10.1111/j.1432-1033.1986.tb09600.x. [DOI] [PubMed] [Google Scholar]
- de Vrij W., Konings W. N. Kinetic characterization of cytochrome c oxidase from Bacillus subtilis. Eur J Biochem. 1987 Aug 3;166(3):581–587. doi: 10.1111/j.1432-1033.1987.tb13553.x. [DOI] [PubMed] [Google Scholar]
- de Vrij W., van den Burg B., Konings W. N. Spectral and potentiometric analysis of cytochromes from Bacillus subtilis. Eur J Biochem. 1987 Aug 3;166(3):589–595. doi: 10.1111/j.1432-1033.1987.tb13554.x. [DOI] [PubMed] [Google Scholar]


