Abstract
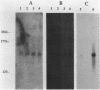
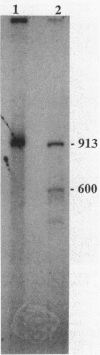
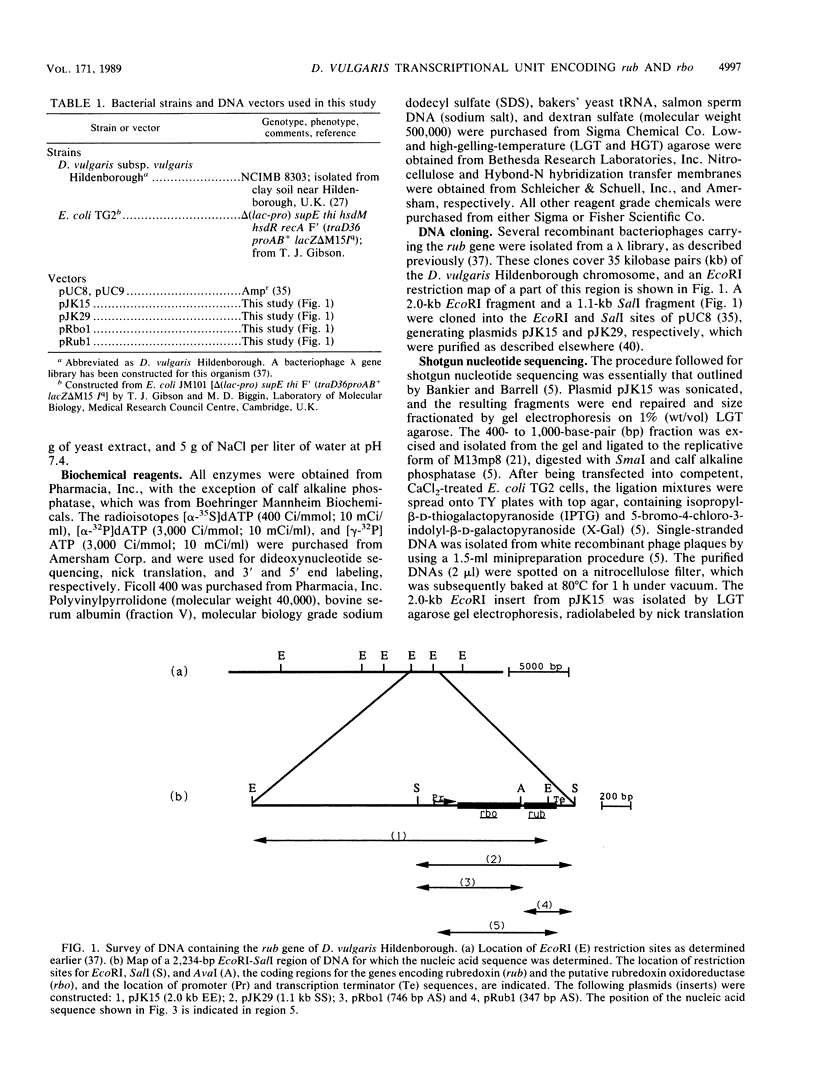
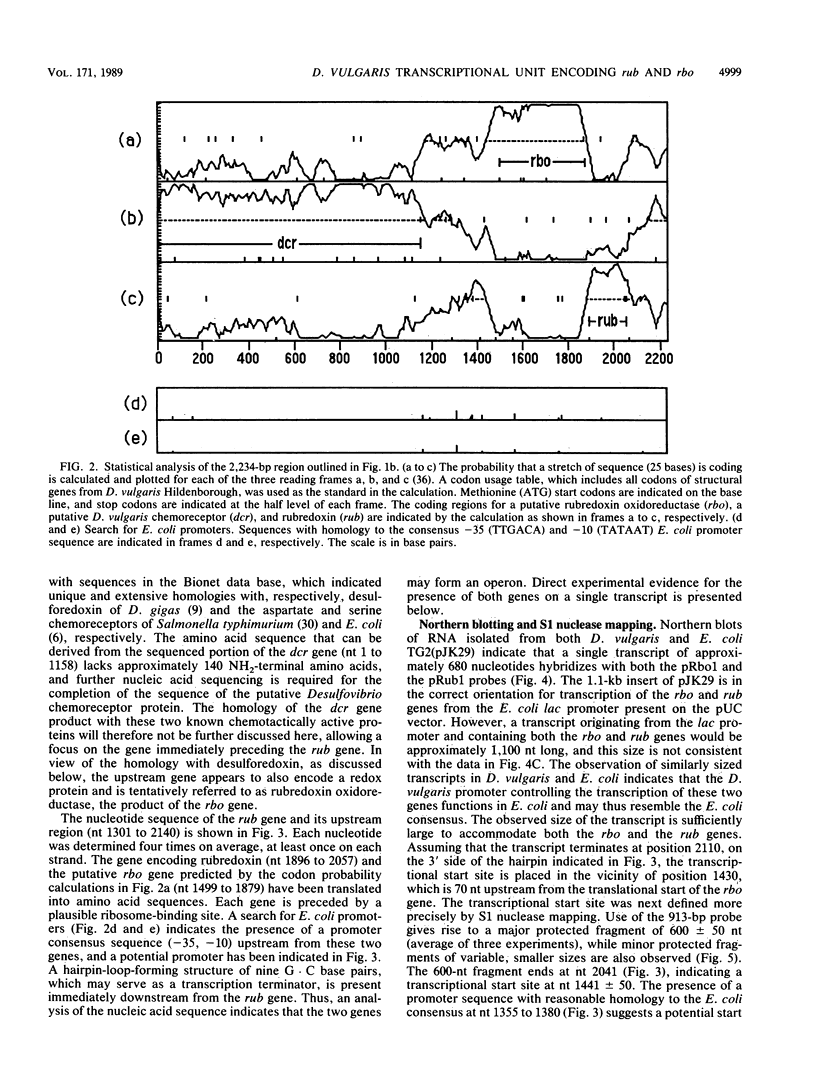
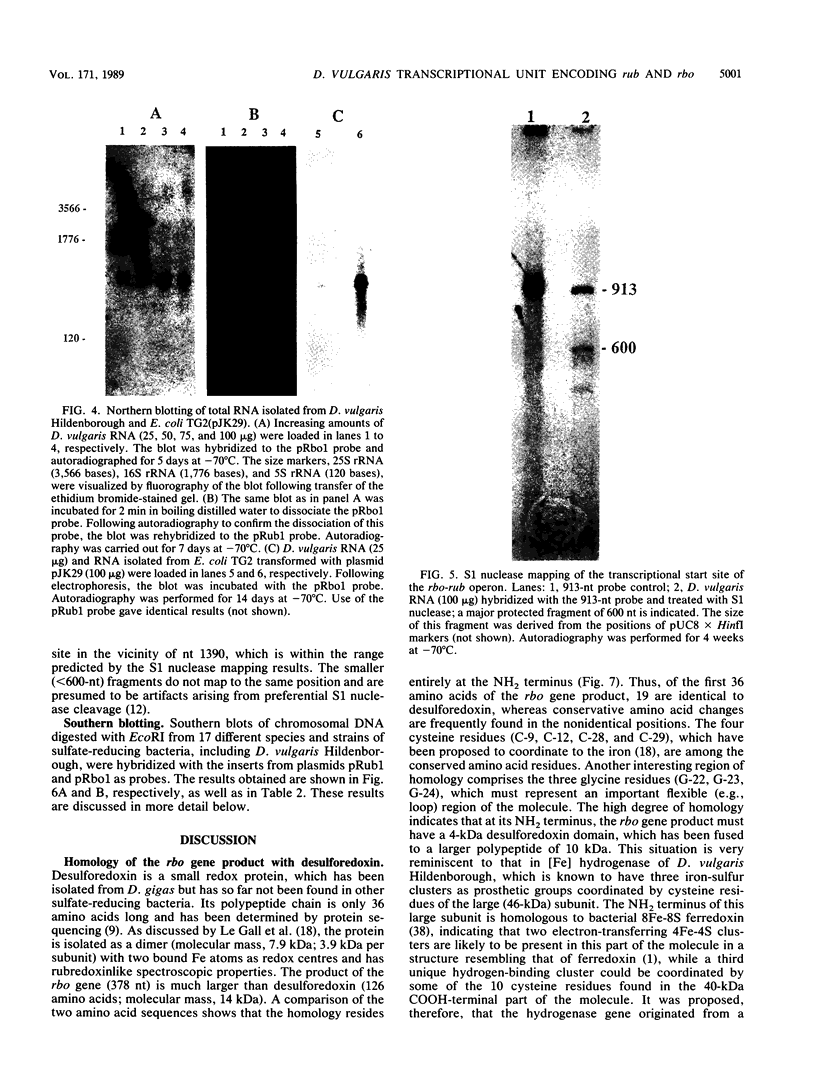
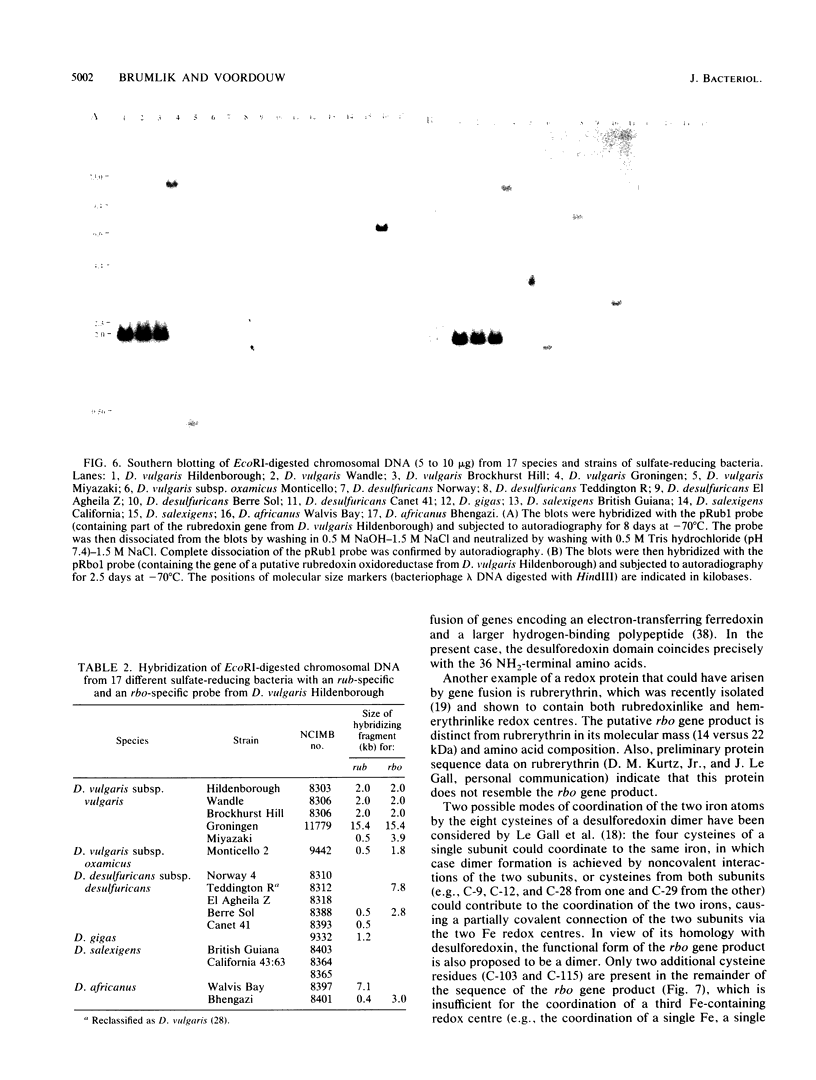
The nucleotide sequence of a 2.0-kilobase-pair EcoRI restriction fragment upstream from the gene (rub, 162 base pairs) encoding rubredoxin from Desulfovibrio vulgaris Hildenborough indicates that it is part of a larger transcriptional unit, containing an additional 378-base-pair open reading frame which terminates 16 nucleotides from the translational start of the rub gene and could encode a polypeptide of 14 kilodaltons (kDa). Northern (RNA) blotting of RNA isolated from both D. vulgaris Hildenborough and Escherichia coli TG2 transformed with plasmid pJK29, which contains both genes on a 1.1-kilobase-pair SalI insert, confirms that the genes for this 14-kDa polypeptide and rubredoxin are present on a single transcript of 680 nucleotides. Strong evidence that the 14-kDa polypeptide is also a redox protein is provided by the fact that its NH2 terminus is homologous to desulforedoxin, which has been isolated from D. gigas as a small dimeric redox protein (36 amino acids per monomer), coordinating two iron atoms. Since rubredoxin is a potential redox partner for the 14-kDa protein, it has been tentatively named rubredoxin oxidoreductase, produced by the rbo gene. Southern blotting indicates that the rbo-rub operon is present in several species and strains of sulfate-reducing bacteria.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adman E. T., Sieker L. C., Jensen L. H., Bruschi M., Le Gall J. A structural model of rubredoxin from Desulfovibrio vulgaris at 2 A resolution. J Mol Biol. 1977 May 5;112(1):113–120. doi: 10.1016/s0022-2836(77)80159-9. [DOI] [PubMed] [Google Scholar]
- Adman E. T., Sieker L. C., Jensen L. H. Structure of a bacterial ferredoxin. J Biol Chem. 1973 Jun 10;248(11):3987–3996. [PubMed] [Google Scholar]
- Bachmayer H., Benson A. M., Yasunobu K. T., Garrard W. T., Whiteley H. R. Nonheme iron proteins. IV. Structural studies of Micrococcus aerogenes rubredoxin. Biochemistry. 1968 Mar;7(3):986–996. doi: 10.1021/bi00843a016. [DOI] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Bruschi M., Moura I., Le Gall J., Xavier A. V., Sieker L. C., Couchoud P. The amino acid sequence of desulforedoxin, a new type of non heme iron protein from Desulfovibrio gigas. Biochem Biophys Res Commun. 1979 Sep 27;90(2):596–605. doi: 10.1016/0006-291x(79)91277-4. [DOI] [PubMed] [Google Scholar]
- Bruschi M. Non-heme iron proteins. The amino acid sequence of rubredoxin from Desulfovibrio vulgaris. Biochim Biophys Acta. 1976 May 20;434(1):4–17. doi: 10.1016/0005-2795(76)90030-1. [DOI] [PubMed] [Google Scholar]
- Bruschi M. The amino acid sequence of rubredoxin from the sulfate reducing bacterium, Desulfovibrio gigas. Biochem Biophys Res Commun. 1976 May 17;70(2):615–621. doi: 10.1016/0006-291x(76)91092-5. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Nakazawa A. Cyclic AMP-dependent initiation and rho-dependent termination of colicin E1 gene transcription. J Biol Chem. 1983 Jun 10;258(11):7072–7078. [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Frey M., Sieker L., Payan F., Haser R., Bruschi M., Pepe G., LeGall J. Rubredoxin from Desulfovibrio gigas. A molecular model of the oxidized form at 1.4 A resolution. J Mol Biol. 1987 Oct 5;197(3):525–541. doi: 10.1016/0022-2836(87)90562-6. [DOI] [PubMed] [Google Scholar]
- Herriott J. R., Sieker L. C., Jensen L. H., Lovenberg W. Structure of rubredoxin: an x-ray study to 2.5 A resolution. J Mol Biol. 1970 Jun 14;50(2):391–406. doi: 10.1016/0022-2836(70)90200-7. [DOI] [PubMed] [Google Scholar]
- Hormel S., Walsh K. A., Prickril B. C., Titani K., LeGall J., Sieker L. C. Amino acid sequence of rubredoxin from Desulfovibrio desulfuricans strain 27774. FEBS Lett. 1986 May 26;201(1):147–150. doi: 10.1016/0014-5793(86)80588-9. [DOI] [PubMed] [Google Scholar]
- Le Gall J. Purification PARTIELLE ET 'ETUDE DE LA NAD: rubrédoxine oxydo-réductase de D. Gigas. Ann Inst Pasteur (Paris) 1968 Jan;114(1):109–115. [PubMed] [Google Scholar]
- LeGall J., Prickril B. C., Moura I., Xavier A. V., Moura J. J., Huynh B. H. Isolation and characterization of rubrerythrin, a non-heme iron protein from Desulfovibrio vulgaris that contains rubredoxin centers and a hemerythrin-like binuclear iron cluster. Biochemistry. 1988 Mar 8;27(5):1636–1642. doi: 10.1021/bi00405a037. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Odom J. M., Peck H. D., Jr Hydrogenase, electron-transfer proteins, and energy coupling in the sulfate-reducing bacteria Desulfovibrio. Annu Rev Microbiol. 1984;38:551–592. doi: 10.1146/annurev.mi.38.100184.003003. [DOI] [PubMed] [Google Scholar]
- Petitdemange H., Marczak R., Blusson H., Gay R. Isolation and properties of reduced nicotinamide adenine dinucleotiderubredoxin oxidoreductase of Clostridium acetobutylicum. Biochem Biophys Res Commun. 1979 Dec 28;91(4):1258–1265. doi: 10.1016/0006-291x(79)91202-6. [DOI] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieker L. C., Stenkamp R. E., Jensen L. H., Prickril B., LeGall J. Structure of rubredoxin from the bacterium Desulfovibrio desulfuricans. FEBS Lett. 1986 Nov 10;208(1):73–76. doi: 10.1016/0014-5793(86)81535-6. [DOI] [PubMed] [Google Scholar]
- Staden R. Graphic methods to determine the function of nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):521–538. doi: 10.1093/nar/12.1part2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R., McLachlan A. D. Codon preference and its use in identifying protein coding regions in long DNA sequences. Nucleic Acids Res. 1982 Jan 11;10(1):141–156. doi: 10.1093/nar/10.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vogel H., Bruschi M., Le Gall J. Phylogenetic studies of two rubredoxins from sulfate reducing bacteria. J Mol Evol. 1977 Apr 29;9(2):111–119. doi: 10.1007/BF01732743. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Brenner S. Nucleotide sequence of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem. 1985 May 2;148(3):515–520. doi: 10.1111/j.1432-1033.1985.tb08869.x. [DOI] [PubMed] [Google Scholar]
- Voordouw G. Cloning of genes encoding redox proteins of known amino acid sequence from a library of the Desulfovibrio vulgaris (Hildenborough) genome. Gene. 1988 Jul 15;67(1):75–83. doi: 10.1016/0378-1119(88)90010-8. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Walker J. E., Brenner S. Cloning of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough) and determination of the NH2-terminal sequence. Eur J Biochem. 1985 May 2;148(3):509–514. doi: 10.1111/j.1432-1033.1985.tb08868.x. [DOI] [PubMed] [Google Scholar]
- WOLIN E. A., WOLIN M. J., WOLFE R. S. FORMATION OF METHANE BY BACTERIAL EXTRACTS. J Biol Chem. 1963 Aug;238:2882–2886. [PubMed] [Google Scholar]
- Watenpaugh K. D., Margulis T. N., Sieker L. C., Jensen L. H. Water structure in a protein crystal: rubredoxin at 1.2 A resolution. J Mol Biol. 1978 Jun 25;122(2):175–190. doi: 10.1016/0022-2836(78)90034-7. [DOI] [PubMed] [Google Scholar]
- Watenpaugh K. D., Sieker L. C., Jensen L. H. Crystallographic refinement of rubredoxin at 1 x 2 A degrees resolution. J Mol Biol. 1980 Apr 15;138(3):615–633. doi: 10.1016/s0022-2836(80)80020-9. [DOI] [PubMed] [Google Scholar]
- Watenpaugh K. D., Sieker L. C., Jensen L. H. The structure of rubredoxin at 1.2 A resolution. J Mol Biol. 1979 Jul 5;131(3):509–522. doi: 10.1016/0022-2836(79)90005-6. [DOI] [PubMed] [Google Scholar]