Abstract
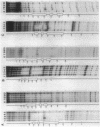
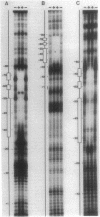
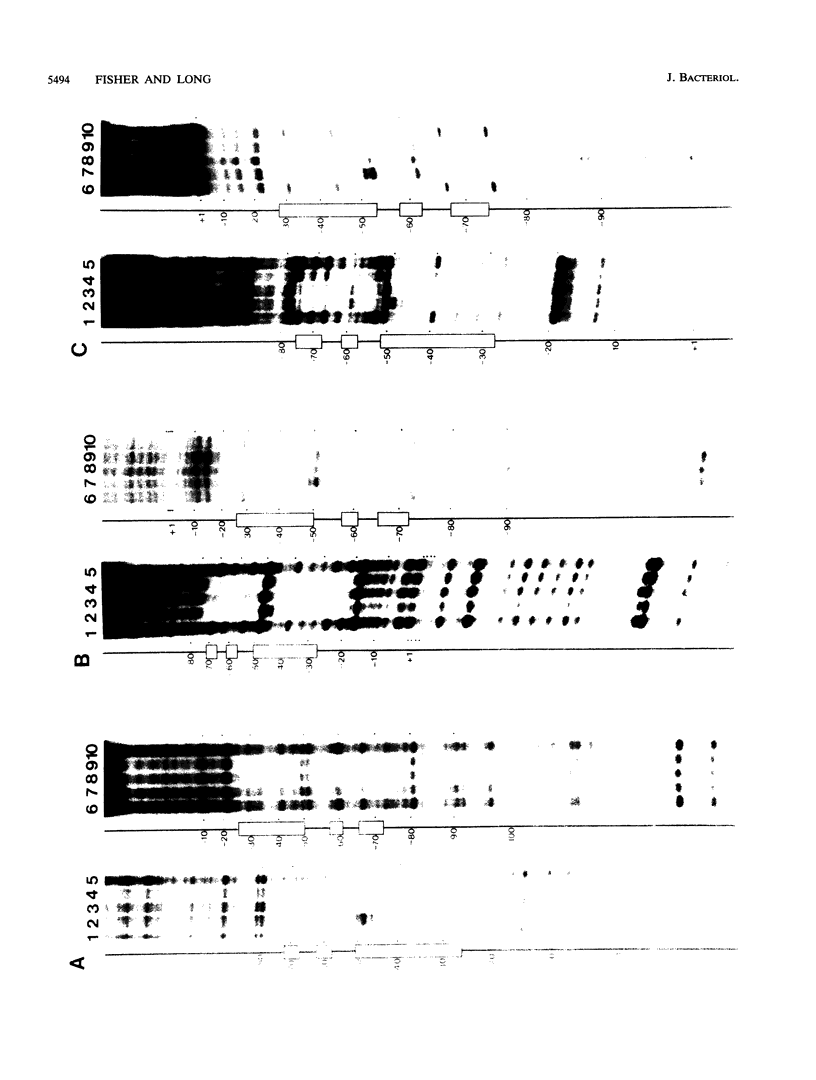
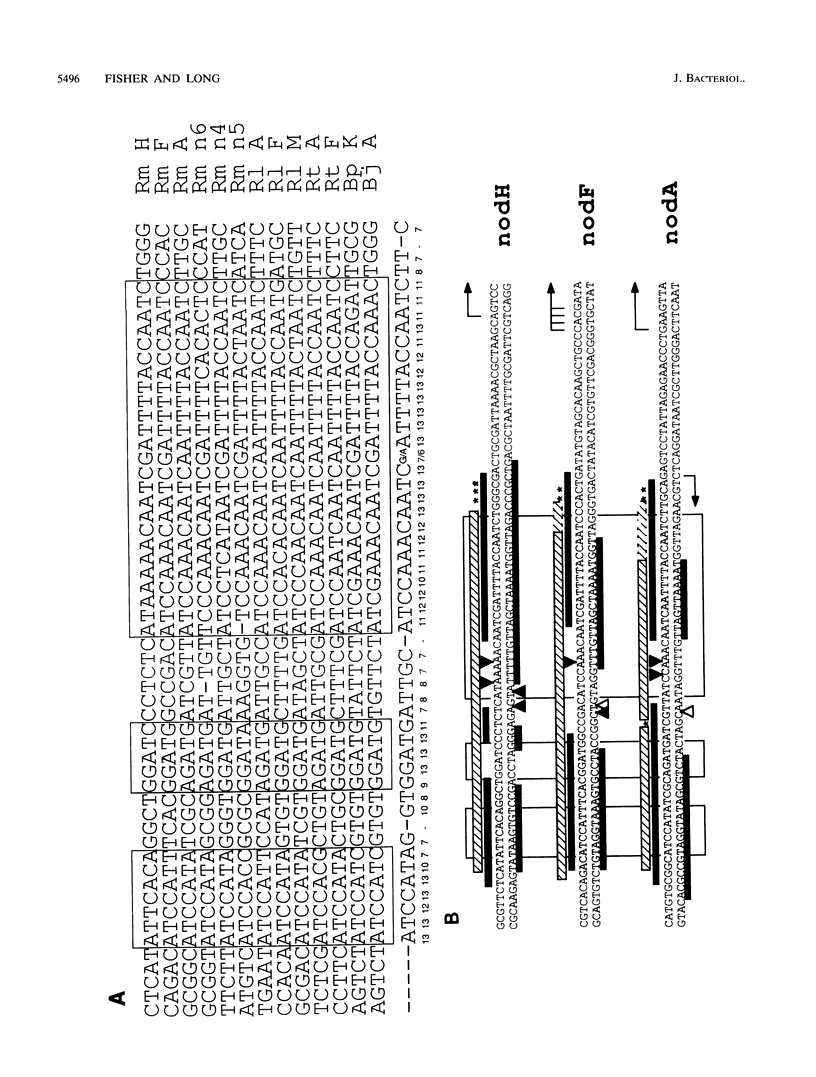
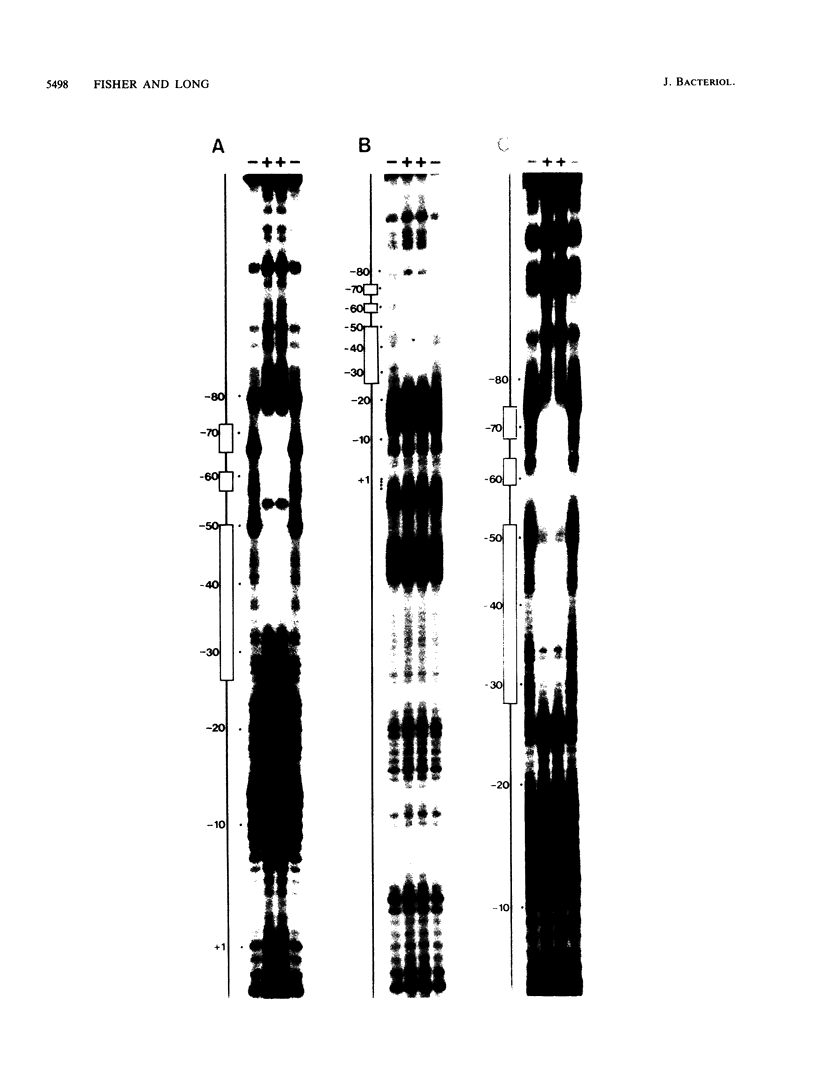
The Rhizobium meliloti nodD1 and nodD3 gene products (NodD1 and NodD3) are members of the lysR-nodD gene regulator family. They are functionally distinct in that NodD1 transcriptionally activates other nod genes in the presence of a flavonoid inducer such as luteolin, while NodD3 is capable of activating nod gene expression at high levels in the absence of inducer. NodD1 and NodD3 are DNA-binding proteins which interact with DNA sequences situated upstream of the transcription initiation sites of at least three sets of inducible nod genes. We report the footprinting of NodD1- and NodD3-DNA complexes with both DNase I and the 1,10-phenanthroline-copper ion reagent. NodD1 and NodD3 both interacted with the nodABC, nodFE, and nodH promoters and protected from cleavage an extensive piece of DNA, including the nod box, from approximately -20 to -75 from the transcription start site for each of the three promoters. The constitutively activating protein NodD3 displayed an additional hypersensitive cleavage site in its footprint compared with NodD1.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi A., Gaillard C., Bernardi G. The specificity of five DNAases as studied by the analysis of 5'-terminal doublets. Eur J Biochem. 1975 Apr 1;52(3):451–457. doi: 10.1111/j.1432-1033.1975.tb04013.x. [DOI] [PubMed] [Google Scholar]
- Bernardi G., Ehrlich S. D., Thiery J. P. The specificity of deoxyribonucleases and their use in nucleotide sequence studies. Nat New Biol. 1973 Nov 14;246(150):36–40. doi: 10.1038/newbio246036a0. [DOI] [PubMed] [Google Scholar]
- Christman M. F., Storz G., Ames B. N. OxyR, a positive regulator of hydrogen peroxide-inducible genes in Escherichia coli and Salmonella typhimurium, is homologous to a family of bacterial regulatory proteins. Proc Natl Acad Sci U S A. 1989 May;86(10):3484–3488. doi: 10.1073/pnas.86.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé F., Rosenberg C., Vasse J., Maillet F., Martinez E., Dénarié J., Truchet G. Assignment of symbiotic developmental phenotypes to common and specific nodulation (nod) genetic loci of Rhizobium meliloti. J Bacteriol. 1986 Dec;168(3):1075–1086. doi: 10.1128/jb.168.3.1075-1086.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debellé F., Sharma S. B. Nucleotide sequence of Rhizobium meliloti RCR2011 genes involved in host specificity of nodulation. Nucleic Acids Res. 1986 Sep 25;14(18):7453–7472. doi: 10.1093/nar/14.18.7453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhoff T. T., Fisher R. F., Jacobs T. W., Mulligan J. T., Long S. R. Nucleotide sequence of Rhizobium meliloti 1021 nodulation genes: nodD is read divergently from nodABC. DNA. 1985 Jun;4(3):241–248. doi: 10.1089/dna.1985.4.241. [DOI] [PubMed] [Google Scholar]
- Egelhoff T. T., Long S. R. Rhizobium meliloti nodulation genes: identification of nodDABC gene products, purification of nodA protein, and expression of nodA in Rhizobium meliloti. J Bacteriol. 1985 Nov;164(2):591–599. doi: 10.1128/jb.164.2.591-599.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Brierley H. L., Mulligan J. T., Long S. R. Transcription of Rhizobium meliloti nodulation genes. Identification of a nodD transcription initiation site in vitro and in vivo. J Biol Chem. 1987 May 15;262(14):6849–6855. [PubMed] [Google Scholar]
- Fisher R. F., Egelhoff T. T., Mulligan J. T., Long S. R. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988 Mar;2(3):282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- Fisher R. F., Swanson J. A., Mulligan J. T., Long S. R. Extended Region of Nodulation Genes in Rhizobium meliloti 1021. II. Nucleotide Sequence, Transcription Start Sites and Protein Products. Genetics. 1987 Oct;117(2):191–201. doi: 10.1093/genetics/117.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Göttfert M., Horvath B., Kondorosi E., Putnoky P., Rodriguez-Quiñones F., Kondorosi A. At least two nodD genes are necessary for efficient nodulation of alfalfa by Rhizobium meliloti. J Mol Biol. 1986 Oct 5;191(3):411–420. doi: 10.1016/0022-2836(86)90136-1. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Haughn G. W., Calvo J. M., Wallace J. C. A large family of bacterial activator proteins. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6602–6606. doi: 10.1073/pnas.85.18.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M. A., Ausubel F. M. Rhizobium meliloti has three functional copies of the nodD symbiotic regulatory gene. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8558–8562. doi: 10.1073/pnas.84.23.8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Bachem C. W., Schell J., Kondorosi A. Host-specific regulation of nodulation genes in Rhizobium is mediated by a plant-signal, interacting with the nodD gene product. EMBO J. 1987 Apr;6(4):841–848. doi: 10.1002/j.1460-2075.1987.tb04829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath B., Kondorosi E., John M., Schmidt J., Török I., Györgypal Z., Barabas I., Wieneke U., Schell J., Kondorosi A. Organization, structure and symbiotic function of Rhizobium meliloti nodulation genes determining host specificity for alfalfa. Cell. 1986 Aug 1;46(3):335–343. doi: 10.1016/0092-8674(86)90654-9. [DOI] [PubMed] [Google Scholar]
- Jacobs T. W., Egelhoff T. T., Long S. R. Physical and genetic map of a Rhizobium meliloti nodulation gene region and nucleotide sequence of nodC. J Bacteriol. 1985 May;162(2):469–476. doi: 10.1128/jb.162.2.469-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondorosi E., Gyuris J., Schmidt J., John M., Duda E., Hoffmann B., Schell J., Kondorosi A. Positive and negative control of nod gene expression in Rhizobium meliloti is required for optimal nodulation. EMBO J. 1989 May;8(5):1331–1340. doi: 10.1002/j.1460-2075.1989.tb03513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara M. D., Sigman D. S. Footprinting DNA-protein complexes in situ following gel retardation assays using 1,10-phenanthroline-copper ion: Escherichia coli RNA polymerase-lac promoter complexes. Biochemistry. 1987 Nov 17;26(23):7234–7238. doi: 10.1021/bi00397a006. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Majors J. Specific binding of CAP factor to lac promoter DNA. Nature. 1975 Aug 21;256(5519):672–674. doi: 10.1038/256672a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller V. L., Taylor R. K., Mekalanos J. J. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987 Jan 30;48(2):271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. A family of activator genes regulates expression of Rhizobium meliloti nodulation genes. Genetics. 1989 May;122(1):7–18. doi: 10.1093/genetics/122.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop A. J., Banfalvi Z., Deshmane N., Gerhold D., Schell M. G., Sirotkin K. M., Stacey G. A locus encoding host range is linked to the common nodulation genes of Bradyrhizobium japonicum. J Bacteriol. 1987 Jun;169(6):2631–2638. doi: 10.1128/jb.169.6.2631-2638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Rossen L., Johnston A. W., Downie J. A. DNA sequence of the Rhizobium leguminosarum nodulation genes nodAB and C required for root hair curling. Nucleic Acids Res. 1984 Dec 21;12(24):9497–9508. doi: 10.1093/nar/12.24.9497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossen L., Shearman C. A., Johnston A. W., Downie J. A. The nodD gene of Rhizobium leguminosarum is autoregulatory and in the presence of plant exudate induces the nodA,B,C genes. EMBO J. 1985 Dec 16;4(13A):3369–3373. doi: 10.1002/j.1460-2075.1985.tb04092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rostas K., Kondorosi E., Horvath B., Simoncsits A., Kondorosi A. Conservation of extended promoter regions of nodulation genes in Rhizobium. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1757–1761. doi: 10.1073/pnas.83.6.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J., John M., Wieneke U., Krüssmann H. D., Schell J. Expression of the nodulation gene nodA in Rhizobium meliloti and localization of the gene product in the cytosol. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9581–9585. doi: 10.1073/pnas.83.24.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield P. R., Watson J. M. DNA sequence of Rhizobium trifolii nodulation genes reveals a reiterated and potentially regulatory sequence preceding nodABC and nodFE. Nucleic Acids Res. 1986 Apr 11;14(7):2891–2903. doi: 10.1093/nar/14.7.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K. F. Conserved nodulation genes from the non-legume symbiont Bradyrhizobium sp. (Parasponia). Nucleic Acids Res. 1986 Apr 11;14(7):2905–2919. doi: 10.1093/nar/14.7.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanblatt S. H., Revzin A. Interactions of the catabolite activator protein (CAP) at the galactose and lactose promoters of Escherichia coli probed by hydroxyl radical footprinting. The second CAP molecule which binds at gal and the one CAP at lac may act to stimulate transcription in the same way. J Biol Chem. 1987 Aug 25;262(24):11422–11427. [PubMed] [Google Scholar]
- Shearman C. A., Rossen L., Johnston A. W., Downie J. A. The Rhizobium leguminosarum nodulation gene nodF encodes a polypeptide similar to acyl-carrier protein and is regulated by nodD plus a factor in pea root exudate. EMBO J. 1986 Apr;5(4):647–652. doi: 10.1002/j.1460-2075.1986.tb04262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A., Tu J. K., Ogawa J., Sanga R., Fisher R. F., Long S. R. Extended Region of Nodulation Genes in Rhizobium meliloti 1021. I. Phenotypes of Tn5 Insertion Mutants. Genetics. 1987 Oct;117(2):181–189. doi: 10.1093/genetics/117.2.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T., O'Neill M., de Crombrugghe B. Interaction site of Escherichia coli cyclic AMP receptor protein on DNA of galactose operon promoters. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5090–5094. doi: 10.1073/pnas.76.10.5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török I., Kondorosi E., Stepkowski T., Pósfai J., Kondorosi A. Nucleotide sequence of Rhizobium meliloti nodulation genes. Nucleic Acids Res. 1984 Dec 21;12(24):9509–9524. doi: 10.1093/nar/12.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wek R. C., Hatfield G. W. Transcriptional activation at adjacent operators in the divergent-overlapping ilvY and ilvC promoters of Escherichia coli. J Mol Biol. 1988 Oct 5;203(3):643–663. doi: 10.1016/0022-2836(88)90199-4. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]