Abstract
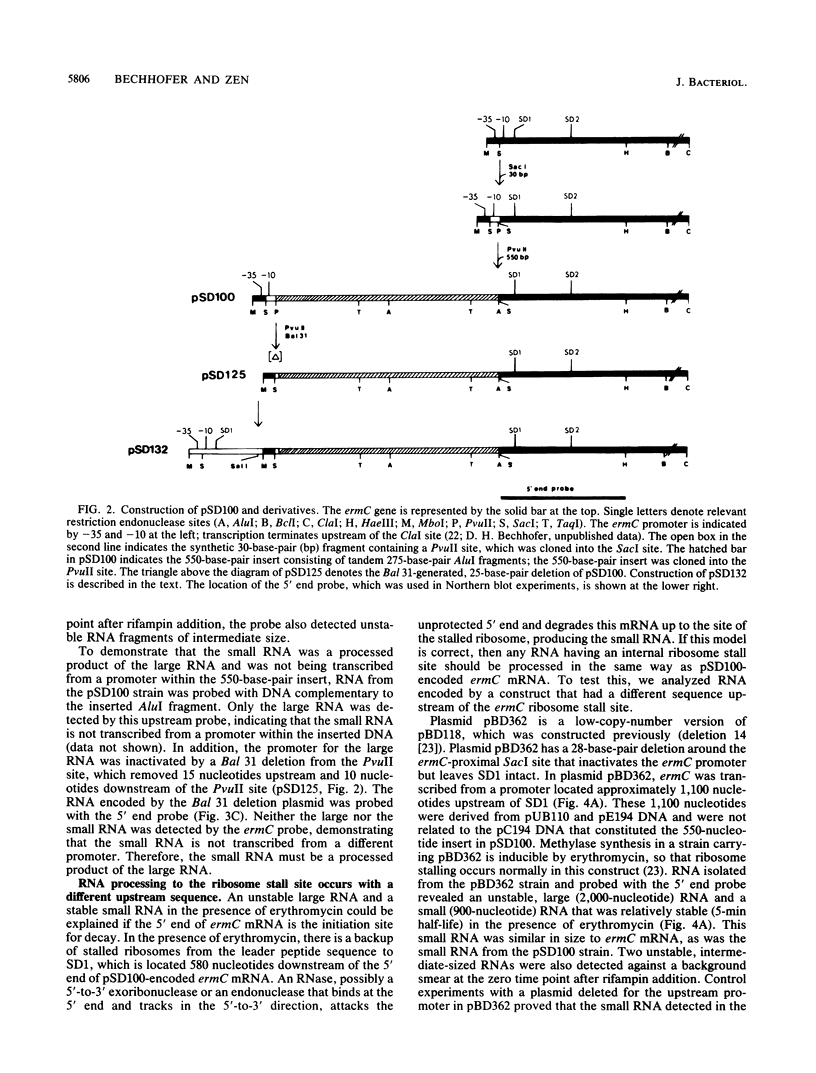
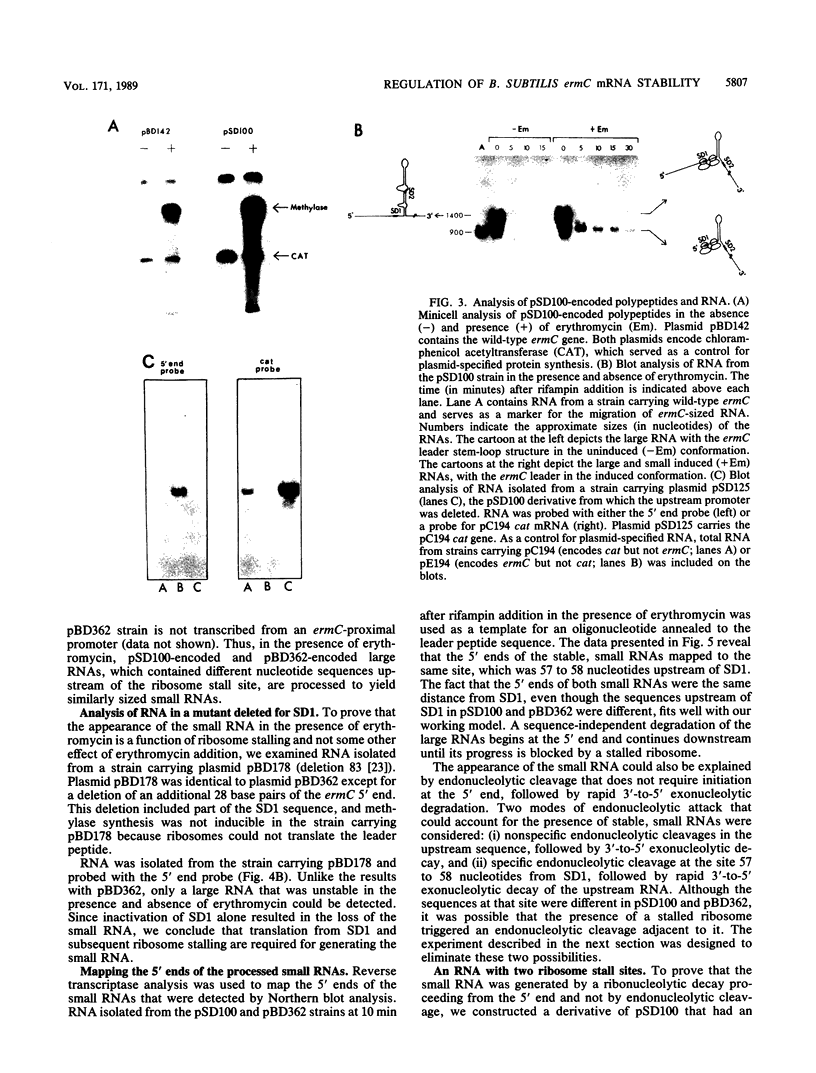
In Bacillus subtilis, the ermC gene encodes an mRNA that is unusually stable (40-min half-life) in the presence of erythromycin, an inducer of ermC gene expression. A requirement for this induced mRNA stability is a ribosome stalled in the ermC leader region. This property of ermC mRNA was used to study the decay of mRNA in B. subtilis. Using constructs in which the ribosome stall site was internal rather than at the 5' end of the message, we show that ribosome stalling provides stability to sequences downstream but not upstream of the ribosome stall site. Our results indicate that ermC mRNA is degraded by a ribonucleolytic activity that begins at the 5' end and degrades the message in a 5'-to-3' direction.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arraiano C. M., Yancey S. D., Kushner S. R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988 Oct;170(10):4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A. 1987 Jan;84(2):498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Nilsson G., von Gabain A., Cohen S. N. The stability of E. coli gene transcripts is dependent on determinants localized to specific mRNA segments. Cell. 1986 Jul 18;46(2):245–251. doi: 10.1016/0092-8674(86)90741-5. [DOI] [PubMed] [Google Scholar]
- Belin D., Mudd E. A., Prentki P., Yi-Yi Y., Krisch H. M. Sense and antisense transcription of bacteriophage T4 gene 32. Processing and stability of the mRNAs. J Mol Biol. 1987 Mar 20;194(2):231–243. doi: 10.1016/0022-2836(87)90371-8. [DOI] [PubMed] [Google Scholar]
- Bothwell A. L., Apirion D. Is RNase V a manifestation of RNase II? Biochem Biophys Res Commun. 1971 Aug 20;44(4):844–851. doi: 10.1016/0006-291x(71)90788-1. [DOI] [PubMed] [Google Scholar]
- Brawerman G. Determinants of messenger RNA stability. Cell. 1987 Jan 16;48(1):5–6. doi: 10.1016/0092-8674(87)90346-1. [DOI] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Cannistraro V. J., Subbarao M. N., Kennell D. Specific endonucleolytic cleavage sites for decay of Escherichia coli mRNA. J Mol Biol. 1986 Nov 20;192(2):257–274. doi: 10.1016/0022-2836(86)90363-3. [DOI] [PubMed] [Google Scholar]
- Castles J. J., Singer M. F. Degradation of polyuridylic acid by ribonuclease II: protection by ribosomes. J Mol Biol. 1969 Feb 28;40(1):1–17. doi: 10.1016/0022-2836(69)90292-7. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Beatty J. T., Cohen S. N., Belasco J. G. An intercistronic stem-loop structure functions as an mRNA decay terminator necessary but insufficient for puf mRNA stability. Cell. 1988 Feb 26;52(4):609–619. doi: 10.1016/0092-8674(88)90473-4. [DOI] [PubMed] [Google Scholar]
- Deutscher M. P. E. coli RNases: making sense of alphabet soup. Cell. 1985 Apr;40(4):731–732. doi: 10.1016/0092-8674(85)90330-7. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan W. P., Kushner S. R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci U S A. 1986 Jan;83(1):120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Dubnau D. Translational attenuation: the regulation of bacterial resistance to the macrolide-lincosamide-streptogramin B antibiotics. CRC Crit Rev Biochem. 1984;16(2):103–132. doi: 10.3109/10409238409102300. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Gorski K., Roch J. M., Prentki P., Krisch H. M. The stability of bacteriophage T4 gene 32 mRNA: a 5' leader sequence that can stabilize mRNA transcripts. Cell. 1985 Dec;43(2 Pt 1):461–469. doi: 10.1016/0092-8674(85)90176-x. [DOI] [PubMed] [Google Scholar]
- Graves M. C., Rabinowitz J. C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for "extended" promoter elements in gram-positive organisms. J Biol Chem. 1986 Aug 25;261(24):11409–11415. [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Hahn J., Contente S., Dubnau D. Replication and incompatibility properties of plasmid pE194 in Bacillus subtilis. J Bacteriol. 1982 Nov;152(2):722–735. doi: 10.1128/jb.152.2.722-735.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J., Grandi G., Gryczan T. J., Dubnau D. Translational attenuation of ermC: a deletion analysis. Mol Gen Genet. 1982;186(2):204–216. doi: 10.1007/BF00331851. [DOI] [PubMed] [Google Scholar]
- Holmes R. K., Singer M. F. Inability to detect RNase V in Escherichia coli and comparison of other ribonucleases before and after infection with coliphage T7. Biochem Biophys Res Commun. 1971 Aug 20;44(4):837–843. doi: 10.1016/0006-291x(71)90787-x. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang C. W., Cantor C. R. Structure of ribosome-bound messenger RNA as revealed by enzymatic accessibility studies. J Mol Biol. 1985 Jan 20;181(2):241–251. doi: 10.1016/0022-2836(85)90088-9. [DOI] [PubMed] [Google Scholar]
- King T. C., Sirdeskmukh R., Schlessinger D. Nucleolytic processing of ribonucleic acid transcripts in procaryotes. Microbiol Rev. 1986 Dec;50(4):428–451. doi: 10.1128/mr.50.4.428-451.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch D. R., Lai M. H. Regulation of a macrolide resistance-beta-galactosidase (ermC-lacZ) gene fusion in Escherichia coli. J Bacteriol. 1984 Jul;159(1):381–384. doi: 10.1128/jb.159.1.381-384.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Kuwano M., Schlessinger D., Apirion D. Ribonuclease V of Escherichia coli. IV. Exonucleolytic cleavage in the 5' to 3' direction with production of 5'-nucleotide monophosphates. J Mol Biol. 1970 Jul 14;51(1):75–82. doi: 10.1016/0022-2836(70)90271-8. [DOI] [PubMed] [Google Scholar]
- Lundberg U., Nilsson G., von Gabain A. The differential stability of the Escherichia coli ompA and bla mRNA at various growth rates is not correlated to the efficiency of translation. Gene. 1988 Dec 10;72(1-2):141–149. doi: 10.1016/0378-1119(88)90136-9. [DOI] [PubMed] [Google Scholar]
- Mayford M., Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989 Mar 5;206(1):69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Mudd E. A., Prentki P., Belin D., Krisch H. M. Processing of unstable bacteriophage T4 gene 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988 Nov;7(11):3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Demonstration of erythromycin-dependent stalling of ribosomes on the ermC leader transcript. J Biol Chem. 1987 Feb 5;262(4):1766–1771. [PubMed] [Google Scholar]
- Narayanan C. S., Dubnau D. Evidence for the translational attenuation model: ribosome-binding studies and structural analysis with an in vitro run-off transcript of ermC. Nucleic Acids Res. 1985 Oct 25;13(20):7307–7326. doi: 10.1093/nar/13.20.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury S. F., Smith N. H., Higgins C. F. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987 Dec 24;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- Nilsson G., Lundberg U., von Gabain A. In vivo and in vitro identity of site specific cleavages in the 5' non-coding region of ompA and bla mRNA in Escherichia coli. EMBO J. 1988 Jul;7(7):2269–2275. doi: 10.1002/j.1460-2075.1988.tb03067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Mendelson N. H., Coyne S. I., Hallock L. L., Cole R. M. Minicells of Bacillus subtilis. J Bacteriol. 1973 May;114(2):860–873. doi: 10.1128/jb.114.2.860-873.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler P., Weisblum B. Erythromycin-induced stabilization of ermA messenger RNA in Staphylococcus aureus and Bacillus subtilis. J Mol Biol. 1988 Oct 20;203(4):905–915. doi: 10.1016/0022-2836(88)90116-7. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Dubnau D. Characterization of a plasmid-specified ribosome methylase associated with macrolide resistance. Nucleic Acids Res. 1981 Jun 11;9(11):2549–2562. doi: 10.1093/nar/9.11.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Dubnau D. Studies on the synthesis of plasmid-coded proteins and their control in Bacillus subtilis minicells. Plasmid. 1979 Apr;2(2):279–289. doi: 10.1016/0147-619x(79)90046-5. [DOI] [PubMed] [Google Scholar]
- Shivakumar A. G., Hahn J., Grandi G., Kozlov Y., Dubnau D. Posttranscriptional regulation of an erythromycin resistance protein specified by plasmic pE194. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3903–3907. doi: 10.1073/pnas.77.7.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz J. A. Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969 Dec 6;224(5223):957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Subbarao M. N., Kennell D. Evidence for endonucleolytic cleavages in decay of lacZ and lacI mRNAs. J Bacteriol. 1988 Jun;170(6):2860–2865. doi: 10.1128/jb.170.6.2860-2865.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakker-Varia S., Ranzini A. C., Dubin D. T. Ribosomal RNA methylation in Staphylococcus aureus and Escherichia coli: effect of the "MLS" (erythromycin resistance) methylase. Plasmid. 1985 Sep;14(2):152–161. doi: 10.1016/0147-619x(85)90075-7. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Siddhikol C., Lai C. J., Demohn V. Erythromycin-inducible resistance in Staphylococcus aureus: requirements for induction. J Bacteriol. 1971 Jun;106(3):835–847. doi: 10.1128/jb.106.3.835-847.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong H. C., Chang S. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc Natl Acad Sci U S A. 1986 May;83(10):3233–3237. doi: 10.1073/pnas.83.10.3233. [DOI] [PMC free article] [PubMed] [Google Scholar]








