Abstract
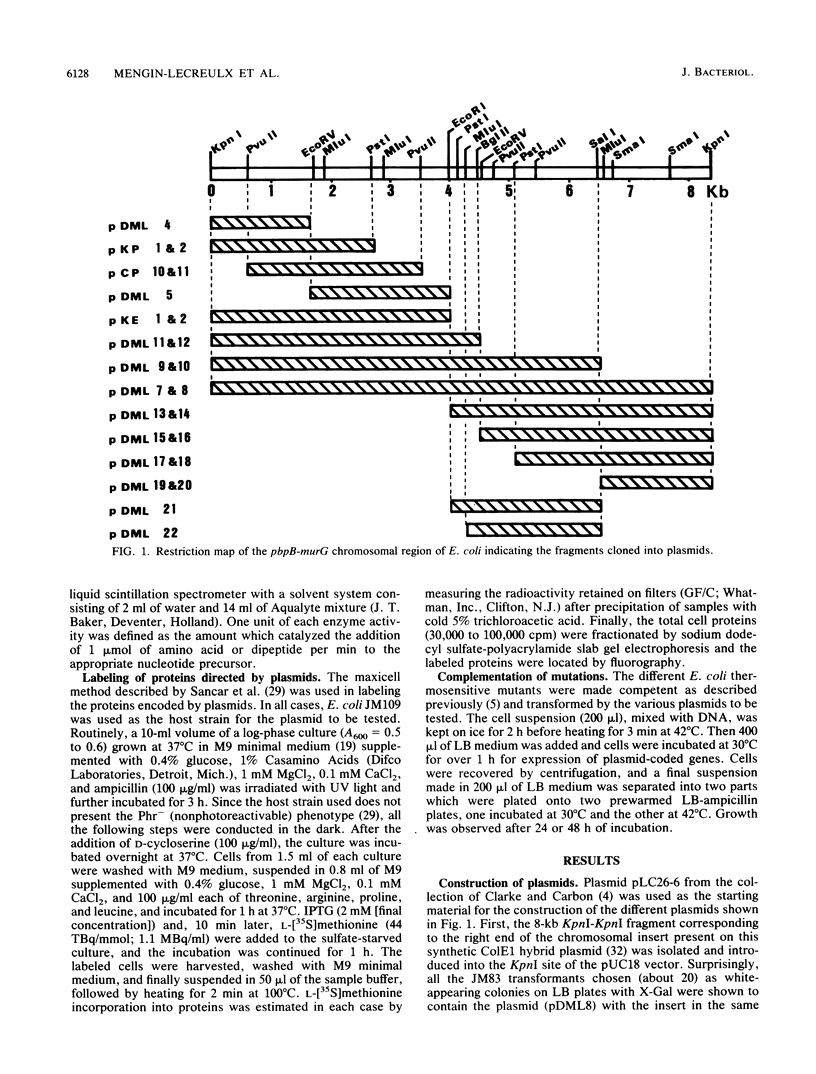
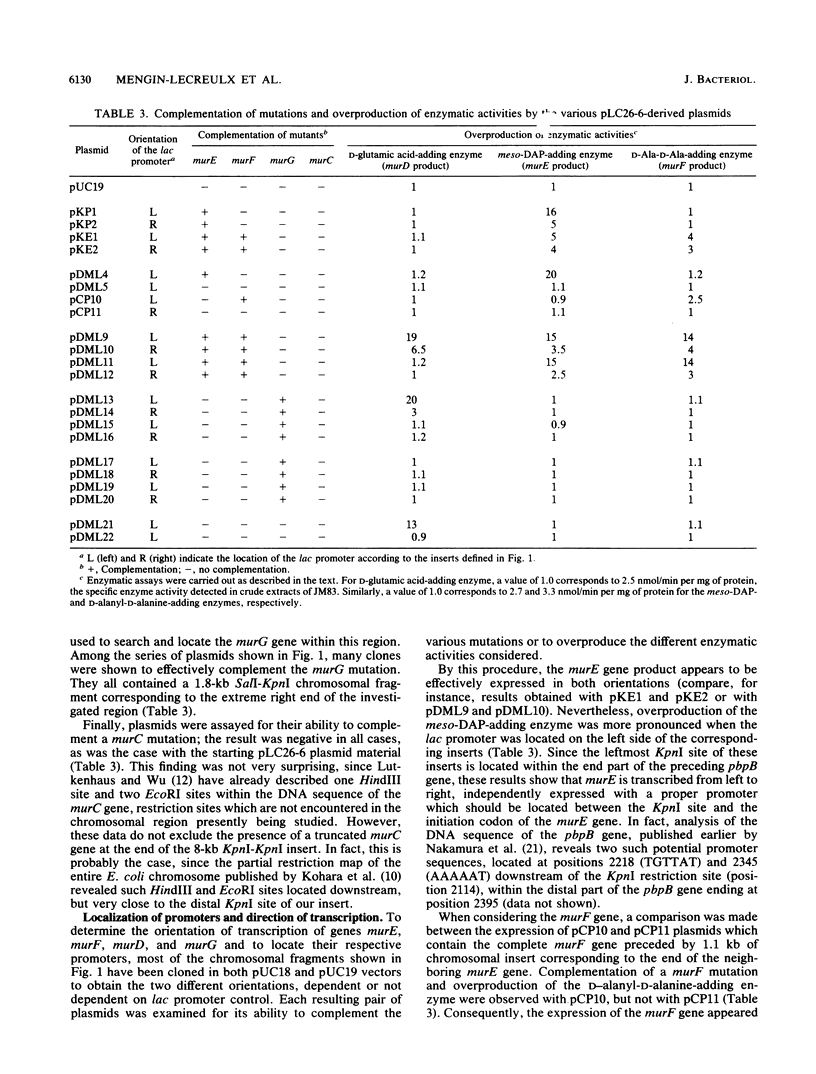
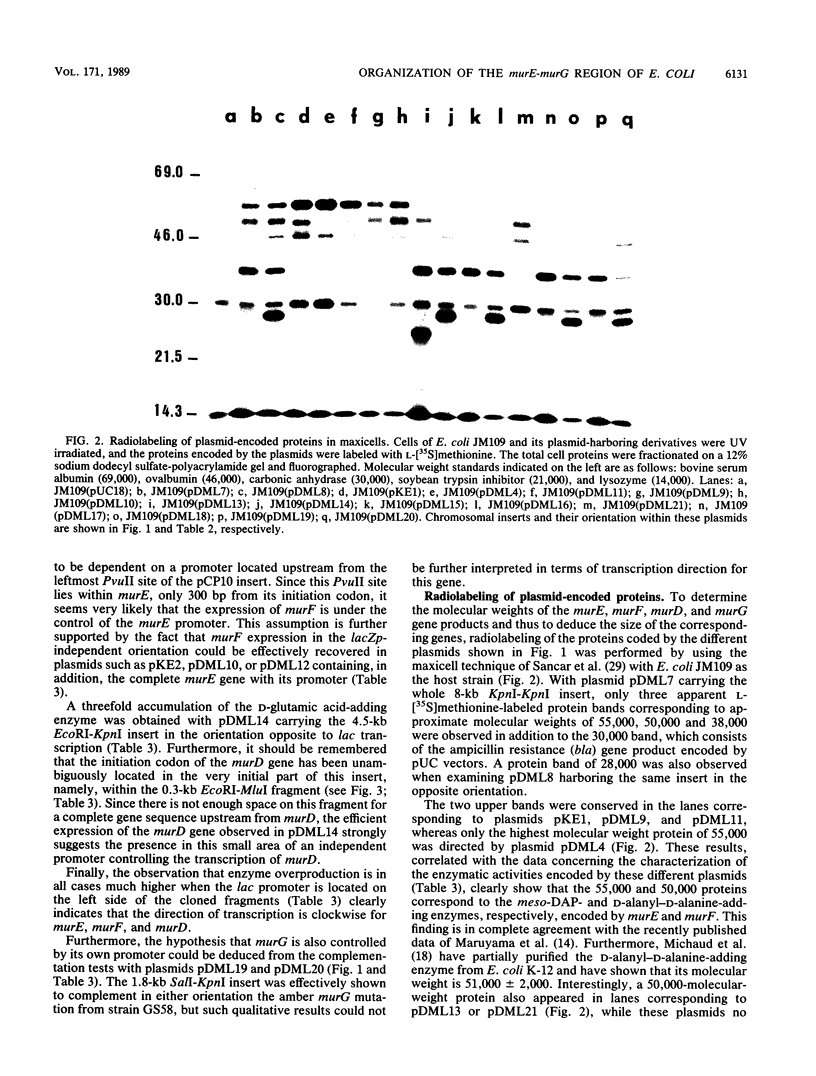
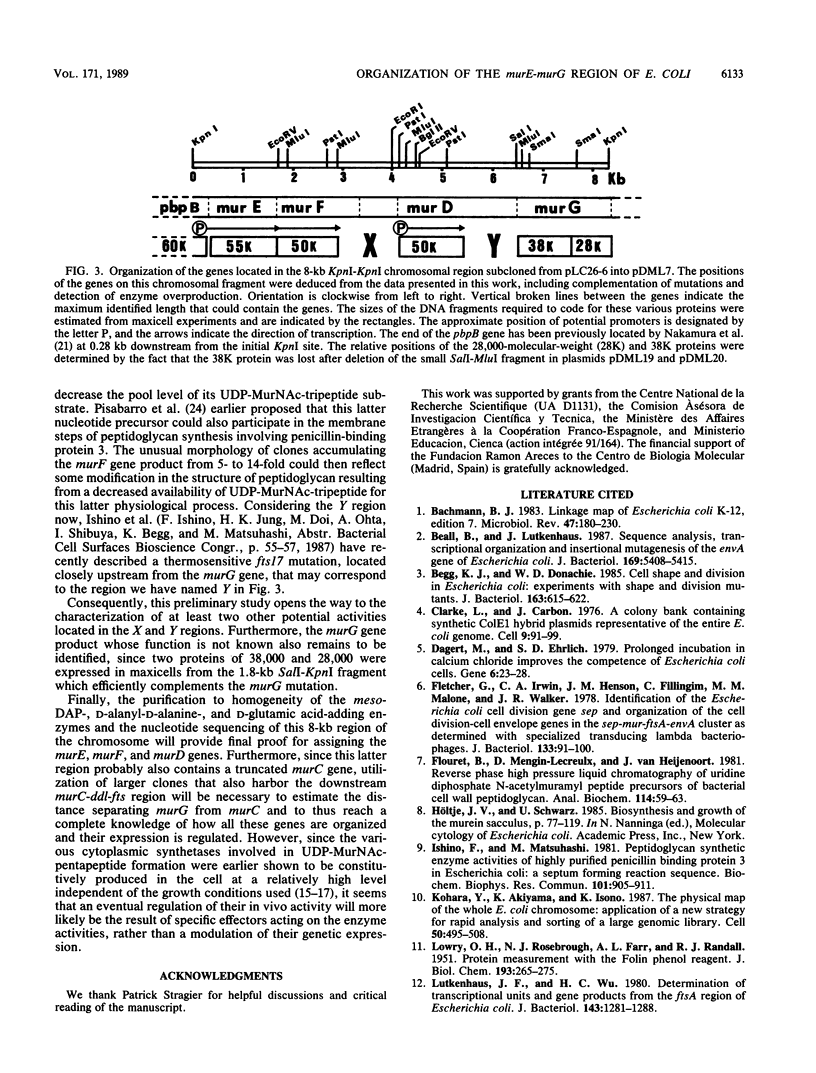
The 2-min region of the Escherichia coli genome contains a large cluster of genes from pbpB to envA that code for proteins involved in peptidoglycan biosynthesis and cell division. From pLC26-6 of the collection of Clarke and Carbon (L. Clarke and J. Carbon, Cell 9:91-99, 1976) plasmids carrying different fragments from the 8-kilobase-pair region downstream of pbpB were constructed and analyzed for their ability to direct protein synthesis in maxicells, to complement various thermosensitive mutations, and to overproduce enzymatic activities. We report the localization of the previously unidentified murD gene coding for the D-glutamic acid-adding enzyme within this region. Our data show that the genes are in the order pbpB-murE-murF-X-murD-Y-murG, where X and Y represent chromosomal fragments from 1 to 1.5 kilobase pairs, possibly coding for unknown proteins. Furthermore, the murE and murF genes, encoding the meso-diaminopimelic acid and D-alanyl-D-alanine-adding enzymes, respectively, may be translationally coupled when transcription is initiated upstream of murE, within the preceding structural gene pbpB coding for penicillin-binding protein 3.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. Sequence analysis, transcriptional organization, and insertional mutagenesis of the envA gene of Escherichia coli. J Bacteriol. 1987 Dec;169(12):5408–5415. doi: 10.1128/jb.169.12.5408-5415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Donachie W. D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985 Aug;163(2):615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L., Carbon J. A colony bank containing synthetic Col El hybrid plasmids representative of the entire E. coli genome. Cell. 1976 Sep;9(1):91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Fletcher G., Irwin C. A., Henson J. M., Fillingim C., Malone M. M., Walker J. R. Identification of the Escherichia coli cell division gene sep and organization of the cell division-cell envelope genes in the sep-mur-ftsA-envA cluster as determined with specialized transducing lambda bacteriophages. J Bacteriol. 1978 Jan;133(1):91–100. doi: 10.1128/jb.133.1.91-100.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouret B., Mengin-Lecreulx D., van Heijenoort J. Reverse-phase high-pressure liquid chromatography of uridine diphosphate N-acetylmuramyl peptide precursors of bacterial cell wall peptidoglycan. Anal Biochem. 1981 Jun;114(1):59–63. doi: 10.1016/0003-2697(81)90451-6. [DOI] [PubMed] [Google Scholar]
- Ishino F., Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981 Aug 14;101(3):905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wu H. C. Determination of transcriptional units and gene products from the ftsA region of Escherichia coli. J Bacteriol. 1980 Sep;143(3):1281–1288. doi: 10.1128/jb.143.3.1281-1288.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama I. N., Yamamoto A. H., Hirota Y. Determination of gene products and coding regions from the murE-murF region of Escherichia coli. J Bacteriol. 1988 Aug;170(8):3786–3788. doi: 10.1128/jb.170.8.3786-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Flouret B., van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982 Sep;151(3):1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Flouret B., van Heijenoort J. Pool levels of UDP N-acetylglucosamine and UDP N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983 Jun;154(3):1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985 Jul;163(1):208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud C., Blanot D., Flouret B., Van Heijenoort J. Partial purification and specificity studies of the D-glutamate-adding and D-alanyl-D-alanine-adding enzymes from Escherichia coli K12. Eur J Biochem. 1987 Aug 3;166(3):631–637. doi: 10.1111/j.1432-1033.1987.tb13560.x. [DOI] [PubMed] [Google Scholar]
- Miyakawa T., Matsuzawa H., Matsuhashi M., Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972 Nov;112(2):950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M., Maruyama I. N., Soma M., Kato J., Suzuki H., Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191(1):1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- Nishimura Y., Takeda Y., Nishimura A., Suzuki H., Inouye M., Hirota Y. Synthetic ColE1 plasmids carrying genes for cell division in Escherichia coli. Plasmid. 1977 Nov;1(1):67–77. doi: 10.1016/0147-619x(77)90009-9. [DOI] [PubMed] [Google Scholar]
- Parquet C., Flouret B., Mengin-Lecreulx D., van Heijenoort J. Nucleotide sequence of the murF gene encoding the UDP-MurNAc-pentapeptide synthetase of Escherichia coli. Nucleic Acids Res. 1989 Jul 11;17(13):5379–5379. doi: 10.1093/nar/17.13.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisabarro A. G., Prats R., Váquez D., Rodríguez-Tébar A. Activity of penicillin-binding protein 3 from Escherichia coli. J Bacteriol. 1986 Oct;168(1):199–206. doi: 10.1128/jb.168.1.199-206.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C., Kenan D. J., Hatfull G. F., Sullivan N. F., Spiegelberg R., Donachie W. D. DNA sequence and transcriptional organization of essential cell division genes ftsQ and ftsA of Escherichia coli: evidence for overlapping transcriptional units. J Bacteriol. 1984 Nov;160(2):546–555. doi: 10.1128/jb.160.2.546-555.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C., Kenan D. J., Sweeney J., Donachie W. D. Further evidence for overlapping transcriptional units in an Escherichia coli cell envelope-cell division gene cluster: DNA sequence and transcriptional organization of the ddl ftsQ region. J Bacteriol. 1986 Sep;167(3):809–817. doi: 10.1128/jb.167.3.809-817.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmond G. P., Lutkenhaus J. F., Donachie W. D. Identification of new genes in a cell envelope-cell division gene cluster of Escherichia coli: cell envelope gene murG. J Bacteriol. 1980 Oct;144(1):438–440. doi: 10.1128/jb.144.1.438-440.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Temperature-sensitive cell division mutants of Escherichia coli with thermolabile penicillin-binding proteins. J Bacteriol. 1977 Jul;131(1):293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan N. F., Donachie W. D. Transcriptional organization within an Escherichia coli cell division gene cluster: direction of transcription of the cell separation gene envA. J Bacteriol. 1984 Nov;160(2):724–732. doi: 10.1128/jb.160.2.724-732.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Nishimura A., Nishimura Y., Yamada M., Yasuda S., Suzuki H., Hirota Y. Synthetic ColE1 Plasmids carrying genes for penicillin-binding proteins in Escherichia coli. Plasmid. 1981 Jul;6(1):86–98. doi: 10.1016/0147-619x(81)90056-1. [DOI] [PubMed] [Google Scholar]
- White R. J. Control of amino sugar metabolism in Escherichia coli and isolation of mutants unable to degrade amino sugars. Biochem J. 1968 Feb;106(4):847–858. doi: 10.1042/bj1060847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman H. J. A genetic map of several mutations affecting the mucopeptide layer of Escherichia coli. Genet Res. 1972 Aug;20(1):65–74. doi: 10.1017/s0016672300013598. [DOI] [PubMed] [Google Scholar]
- Wijsman H. J. The characterization of an alanine racemase mutant of Escherichia coli. Genet Res. 1972 Dec;20(3):269–277. doi: 10.1017/s001667230001380x. [DOI] [PubMed] [Google Scholar]
- Wild J., Hennig J., Lobocka M., Walczak W., Kłopotowski T. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol Gen Genet. 1985;198(2):315–322. doi: 10.1007/BF00383013. [DOI] [PubMed] [Google Scholar]
- Wu H. C., Venkateswaran P. S. Fosfomycin-resistant mutant of Escherichia coli. Ann N Y Acad Sci. 1974 May 10;235(0):587–592. doi: 10.1111/j.1749-6632.1974.tb43292.x. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]




