Abstract
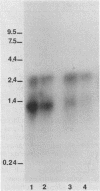
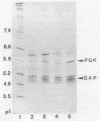
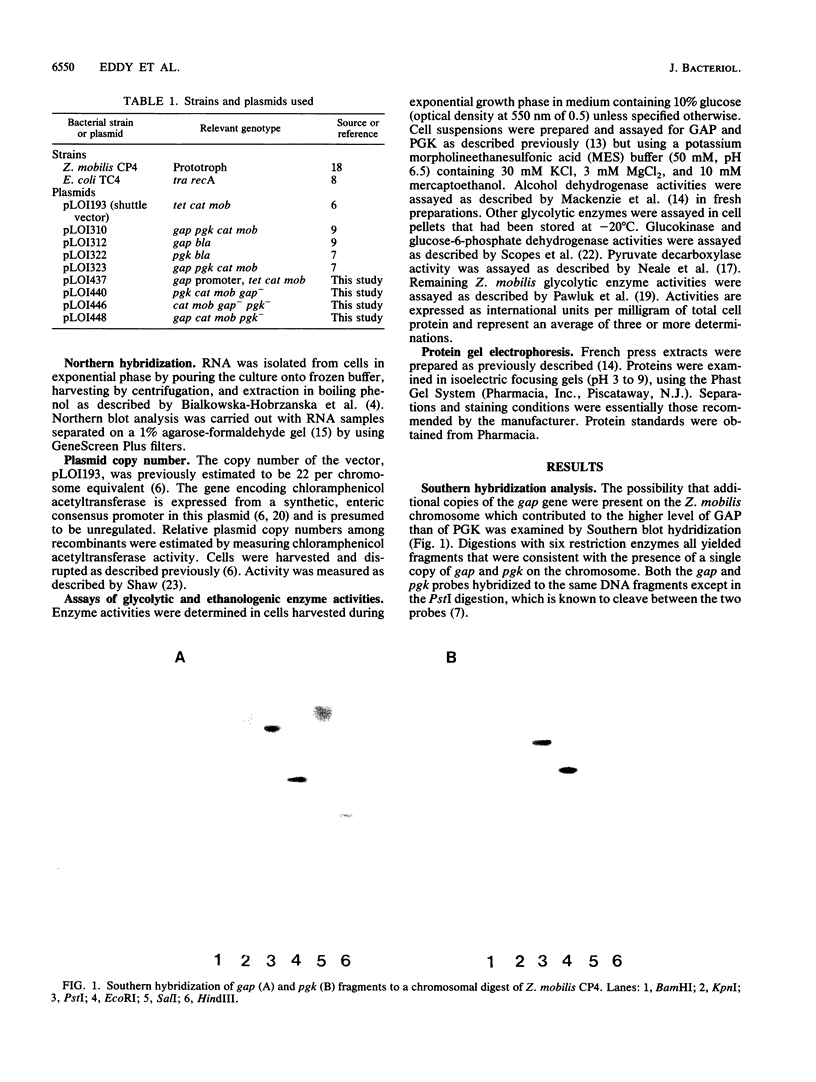
In Zymomonas mobilis, the genes encoding glyceraldehyde-3-phosphate dehydrogenase (GAP) and phosphoglycerate kinase (PGK) are encoded in an operon that is transcribed from tandem promoters. The promoter-proximal gap gene is expressed at six- to ninefold higher levels than the pgk gene from chromosomal genes and from multiple copies of plasmid-borne genes. Two dominant transcripts were identified. The smaller, most abundant transcript contained primarily the gap message, whereas the larger, less abundant message contained both genes. The ratio of message levels for gap and pgk was calculated to be 5:1 and is sufficient to account for the observed differences in levels of GAP and PGK. The differences in message abundance are proposed to result from either transcriptional attenuation or preferential degradation of the 3' region encoding pgk. Increases in gene dosage were accompanied by one-third the expected increase in enzymatic activity on the basis of estimates of copy number, consistent with the presence of a limiting, positive regulatory factor. However, GAP and PGK expressions were not reduced from the chromosome in recombinants that contained multiple copies of the gap operon with inactive genes.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear D. G., Peabody D. S. The E. coli Rho protein: an ATPase that terminates transcription. Trends Biochem Sci. 1988 Sep;13(9):343–347. doi: 10.1016/0968-0004(88)90104-1. [DOI] [PubMed] [Google Scholar]
- Belasco J. G., Beatty J. T., Adams C. W., von Gabain A., Cohen S. N. Differential expression of photosynthesis genes in R. capsulata results from segmental differences in stability within the polycistronic rxcA transcript. Cell. 1985 Jan;40(1):171–181. doi: 10.1016/0092-8674(85)90320-4. [DOI] [PubMed] [Google Scholar]
- Bialkowska-Hobrzanska H., Gilchrist C. A., Denhardt D. T. Escherichia coli rep gene: identification of the promoter and N terminus of the rep protein. J Bacteriol. 1985 Dec;164(3):1004–1010. doi: 10.1128/jb.164.3.1004-1010.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Byun M. O., Ingram L. O. Expression Vector for Zymomonas mobilis. Appl Environ Microbiol. 1987 Feb;53(2):235–241. doi: 10.1128/aem.53.2.235-241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Ingram L. O. Gene expression in Zymomonas mobilis: promoter structure and identification of membrane anchor sequences forming functional lacZ' fusion proteins. J Bacteriol. 1987 Jun;169(6):2327–2335. doi: 10.1128/jb.169.6.2327-2335.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland M. J., Holland J. P. Isolation and identification of yeast messenger ribonucleic acids coding for enolase, glyceraldehyde-3-phosphate dehydrogenase, and phosphoglycerate kinase. Biochemistry. 1978 Nov 14;17(23):4900–4907. doi: 10.1021/bi00616a007. [DOI] [PubMed] [Google Scholar]
- Mackenzie K. F., Conway T., Aldrich H. C., Ingram L. O. Expression of Zymomonas mobilis adhB (encoding alcohol dehydrogenase II) and adhB-lacZ operon fusions in recombinant Z. mobilis. J Bacteriol. 1989 Sep;171(9):4577–4582. doi: 10.1128/jb.171.9.4577-4582.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie K. F., Eddy C. K., Ingram L. O. Modulation of alcohol dehydrogenase isoenzyme levels in Zymomonas mobilis by iron and zinc. J Bacteriol. 1989 Feb;171(2):1063–1067. doi: 10.1128/jb.171.2.1063-1067.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale A. D., Scopes R. K., Wettenhall R. E., Hoogenraad N. J. Nucleotide sequence of the pyruvate decarboxylase gene from Zymomonas mobilis. Nucleic Acids Res. 1987 Feb 25;15(4):1753–1761. doi: 10.1093/nar/15.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Scopes R. K., Griffiths-Smith K. Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from glyceraldehyde-3-phosphate dehydrogenase through to pyruvate kinase. Biochem J. 1986 Aug 15;238(1):275–281. doi: 10.1042/bj2380275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J. J., Soberon X., Marumoto Y., McMahon J., Itakura K. Biological expression of an Escherichia coli consensus sequence promoter and some mutant derivatives. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3203–3207. doi: 10.1073/pnas.80.11.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K., Testolin V., Stoter A., Griffiths-Smith K., Algar E. M. Simultaneous purification and characterization of glucokinase, fructokinase and glucose-6-phosphate dehydrogenase from Zymomonas mobilis. Biochem J. 1985 Jun 15;228(3):627–634. doi: 10.1042/bj2280627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Youvan D. C., Bylina E. J., Alberti M., Begusch H., Hearst J. E. Nucleotide and deduced polypeptide sequences of the photosynthetic reaction-center, B870 antenna, and flanking polypeptides from R. capsulata. Cell. 1984 Jul;37(3):949–957. doi: 10.1016/0092-8674(84)90429-x. [DOI] [PubMed] [Google Scholar]