Abstract
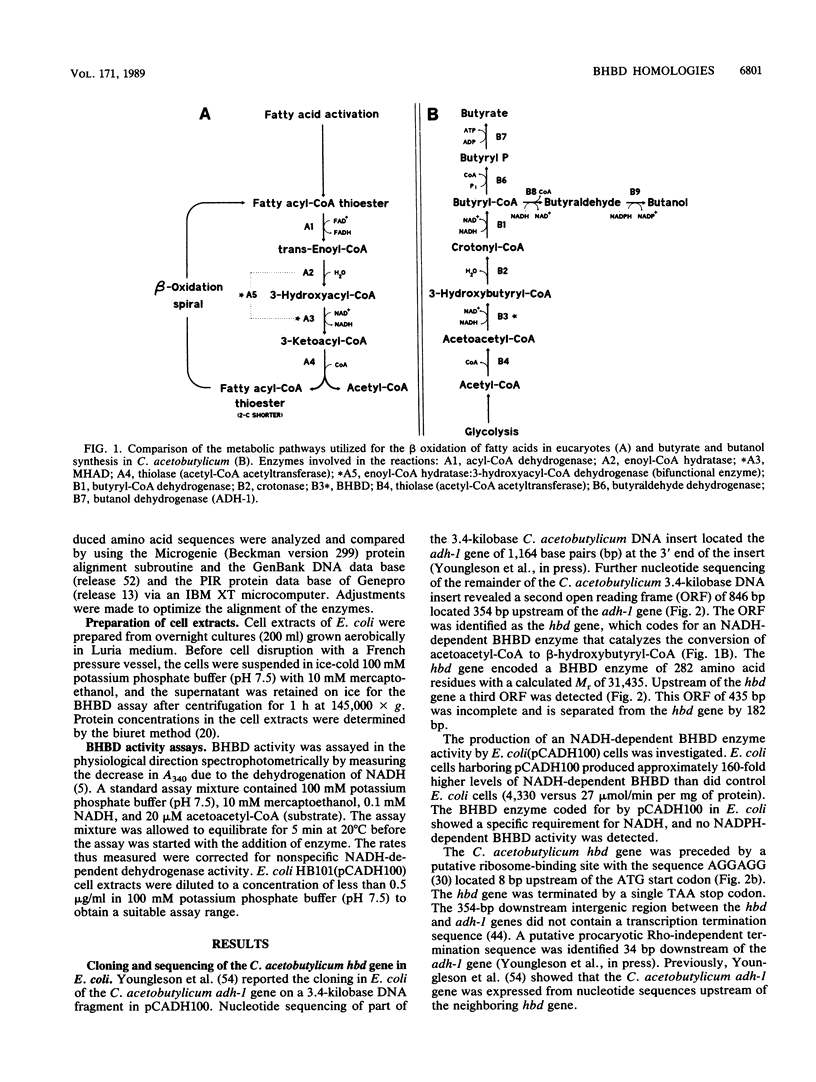
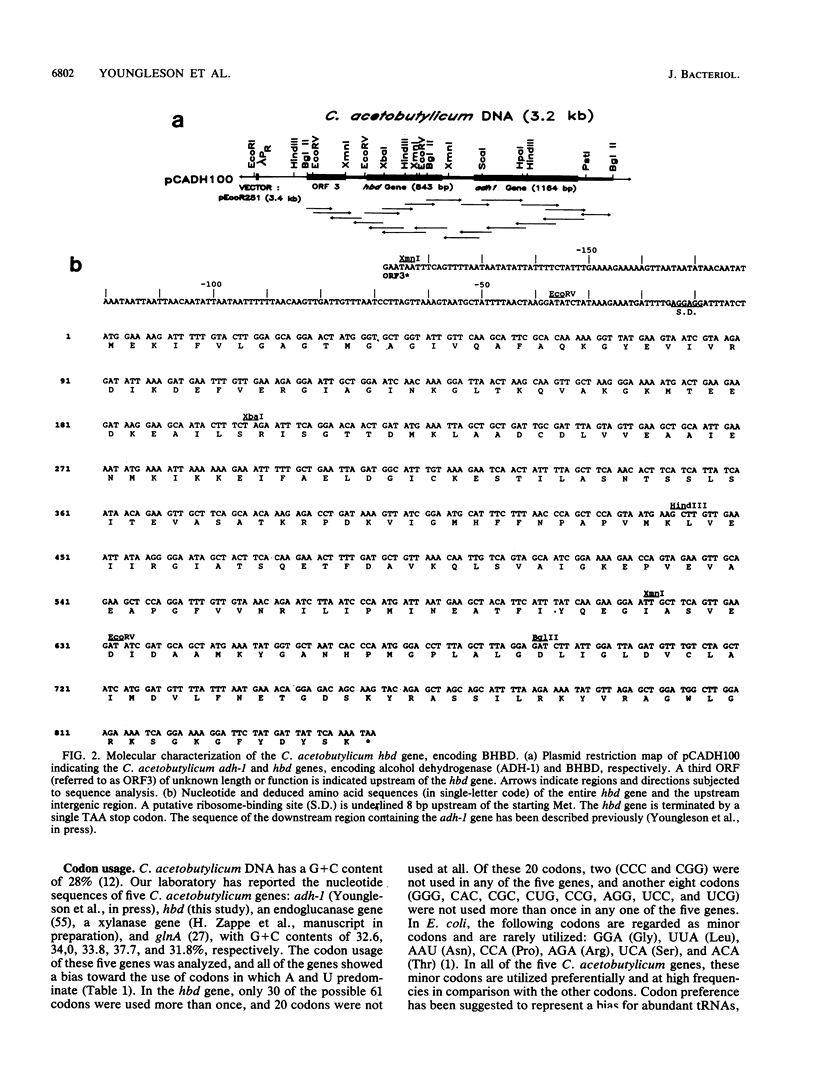
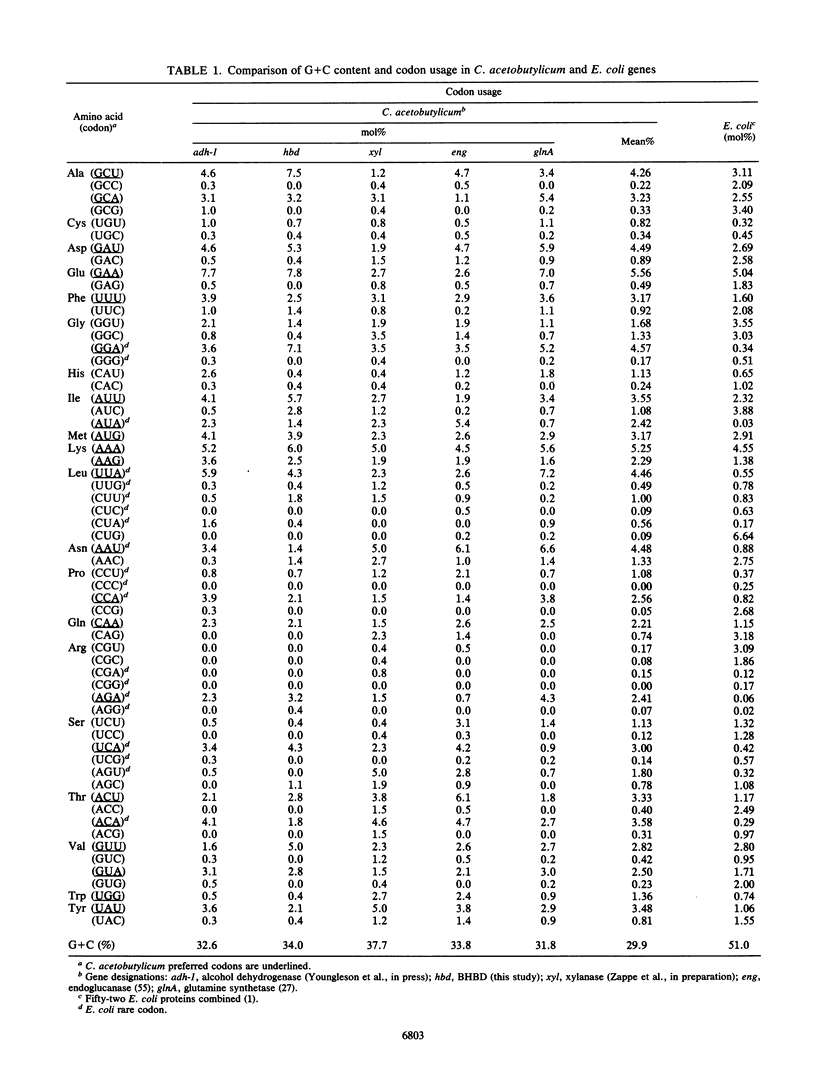
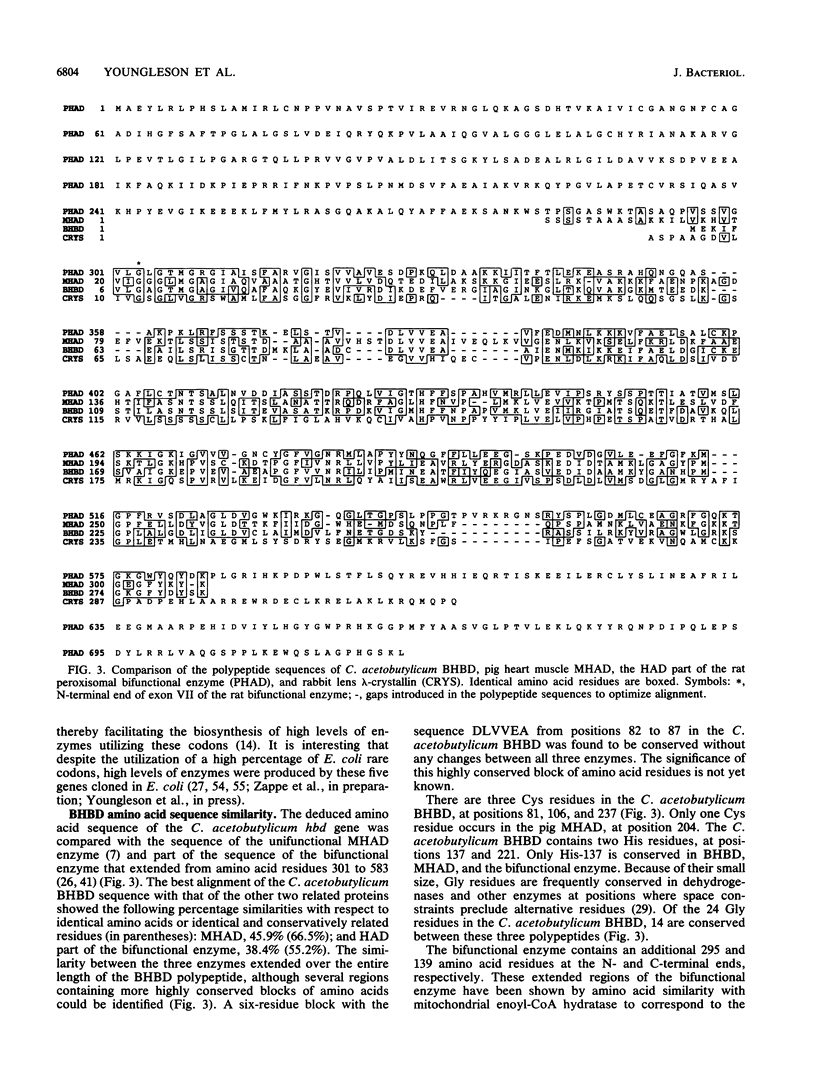
The enzymes NAD-dependent beta-hydroxybutyryl coenzyme A dehydrogenase (BHBD) and 3-hydroxyacetyl coenzyme A (3-hydroxyacyl-CoA) dehydrogenase are part of the central fermentation pathways for butyrate and butanol production in the gram-positive anaerobic bacterium Clostridium acetobutylicum and for the beta oxidation of fatty acids in eucaryotes, respectively. The C. acetobutylicum hbd gene encoding a bacterial BHBD was cloned, expressed, and sequenced in Escherichia coli. The deduced primary amino acid sequence of the C. acetobutylicum BHBD showed 45.9% similarity with the equivalent mitochondrial fatty acid beta-oxidation enzyme and 38.4% similarity with the 3-hydroxyacyl-CoA dehydrogenase part of the bifunctional enoyl-CoA hydratase:3-hydroxyacyl-CoA dehydrogenase from rat peroxisomes. The pig mitochondrial 3-hydroxyacyl-CoA dehydrogenase showed 31.7% similarity with the 3-hydroxyacyl-CoA dehydrogenase part of the bifunctional enzyme from rat peroxisomes. The phylogenetic relationship between these enzymes supports a common evolutionary origin for the fatty acid beta-oxidation pathways of vertebrate mitochondria and peroxisomes and the bacterial fermentation pathway.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alff-Steinberger C. Evidence for a coding pattern on the non-coding strand of the E. coli genome. Nucleic Acids Res. 1984 Mar 12;12(5):2235–2241. doi: 10.1093/nar/12.5.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaya Y., Takiguchi M., Hashimoto T., Mori M. Molecular cloning of cDNA for rat mitochondrial 3-hydroxyacyl-CoA dehydrogenase. Eur J Biochem. 1986 Apr 1;156(1):9–14. doi: 10.1111/j.1432-1033.1986.tb09541.x. [DOI] [PubMed] [Google Scholar]
- Binstock J. F., Schulz H. Fatty acid oxidation complex from Escherichia coli. Methods Enzymol. 1981;71(Pt 100):403–411. doi: 10.1016/0076-6879(81)71051-6. [DOI] [PubMed] [Google Scholar]
- Birktoft J. J., Holden H. M., Hamlin R., Xuong N. H., Banaszak L. J. Structure of L-3-hydroxyacyl-coenzyme A dehydrogenase: preliminary chain tracing at 2.8-A resolution. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8262–8266. doi: 10.1073/pnas.84.23.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitar K. G., Perez-Aranda A., Bradshaw R. A. Amino acid sequence of L-3-hydroxyacyl CoA dehydrogenase from pig heart muscle. FEBS Lett. 1980 Jul 28;116(2):196–198. doi: 10.1016/0014-5793(80)80642-9. [DOI] [PubMed] [Google Scholar]
- Borst P. How proteins get into microbodies (peroxisomes, glyoxysomes, glycosomes). Biochim Biophys Acta. 1986 May 5;866(4):179–203. doi: 10.1016/0167-4781(86)90044-8. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. The simultaneous symbiotic origin of mitochondria, chloroplasts, and microbodies. Ann N Y Acad Sci. 1987;503:55–71. doi: 10.1111/j.1749-6632.1987.tb40597.x. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Clewell D. B. Nature of Col E 1 plasmid replication in Escherichia coli in the presence of the chloramphenicol. J Bacteriol. 1972 May;110(2):667–676. doi: 10.1128/jb.110.2.667-676.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Eventoff W., Rossmann M. G. The evolution of dehydrogenases and kinases. CRC Crit Rev Biochem. 1975 Aug;3(2):111–140. doi: 10.3109/10409237509102554. [DOI] [PubMed] [Google Scholar]
- Fitch W. M., Margoliash E. Construction of phylogenetic trees. Science. 1967 Jan 20;155(3760):279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- Garnier J., Osguthorpe D. J., Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978 Mar 25;120(1):97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Gascuel O., Golmard J. L. A simple method for predicting the secondary structure of globular proteins: implications and accuracy. Comput Appl Biosci. 1988 Aug;4(3):357–365. doi: 10.1093/bioinformatics/4.3.357. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Doolittle W. F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982 Mar;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmanis M. G., Gatenbeck S. Intermediary Metabolism in Clostridium acetobutylicum: Levels of Enzymes Involved in the Formation of Acetate and Butyrate. Appl Environ Microbiol. 1984 Jun;47(6):1277–1283. doi: 10.1128/aem.47.6.1277-1283.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto T. Individual peroxisomal beta-oxidation enzymes. Ann N Y Acad Sci. 1982;386:5–12. doi: 10.1111/j.1749-6632.1982.tb21403.x. [DOI] [PubMed] [Google Scholar]
- Hawkins J. D. A survey on intron and exon lengths. Nucleic Acids Res. 1988 Nov 11;16(21):9893–9908. doi: 10.1093/nar/16.21.9893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Ishii N., Hijikata M., Osumi T., Hashimoto T. Structural organization of the gene for rat enoyl-CoA hydratase:3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme. J Biol Chem. 1987 Jun 15;262(17):8144–8150. [PubMed] [Google Scholar]
- Janssen P. J., Jones W. A., Jones D. T., Woods D. R. Molecular analysis and regulation of the glnA gene of the gram-positive anaerobe Clostridium acetobutylicum. J Bacteriol. 1988 Jan;170(1):400–408. doi: 10.1128/jb.170.1.400-408.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Woods D. R. Acetone-butanol fermentation revisited. Microbiol Rev. 1986 Dec;50(4):484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H., Persson B., Jeffery J. Characteristics of alcohol/polyol dehydrogenases. The zinc-containing long-chain alcohol dehydrogenases. Eur J Biochem. 1987 Sep 1;167(2):195–201. doi: 10.1111/j.1432-1033.1987.tb13323.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lazarow P. B., De Duve C. A fatty acyl-CoA oxidizing system in rat liver peroxisomes; enhancement by clofibrate, a hypolipidemic drug. Proc Natl Acad Sci U S A. 1976 Jun;73(6):2043–2046. doi: 10.1073/pnas.73.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow P. B., Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Misra T. K. A new strategy to create ordered deletions for rapid nucleotide sequencing. Gene. 1985;34(2-3):263–268. doi: 10.1016/0378-1119(85)90135-0. [DOI] [PubMed] [Google Scholar]
- Moreno de la Garza M., Schultz-Borchard U., Crabb J. W., Kunau W. H. Peroxisomal beta-oxidation system of Candida tropicalis. Purification of a multifunctional protein possessing enoyl-CoA hydratase, 3-hydroxyacyl-CoA dehydrogenase and 3-hydroxyacyl-CoA epimerase activities. Eur J Biochem. 1985 Apr 15;148(2):285–291. doi: 10.1111/j.1432-1033.1985.tb08837.x. [DOI] [PubMed] [Google Scholar]
- Mulders J. W., Hendriks W., Blankesteijn W. M., Bloemendal H., de Jong W. W. Lambda-crystallin, a major rabbit lens protein, is related to hydroxyacyl-coenzyme A dehydrogenases. J Biol Chem. 1988 Oct 25;263(30):15462–15466. [PubMed] [Google Scholar]
- Naora H., Deacon N. J. Relationship between the total size of exons and introns in protein-coding genes of higher eukaryotes. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6196–6200. doi: 10.1073/pnas.79.20.6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttley W. M., Aitchison J. D., Rachubinski R. A. cDNA cloning and primary structure determination of the peroxisomal trifunctional enzyme hydratase-dehydrogenase-epimerase from the yeast Candida tropicalis pK233. Gene. 1988 Sep 30;69(2):171–180. doi: 10.1016/0378-1119(88)90428-3. [DOI] [PubMed] [Google Scholar]
- Opperdoes F. R. Glycosomes may provide clues to the import of peroxisomal proteins. Trends Biochem Sci. 1988 Jul;13(7):255–260. doi: 10.1016/0968-0004(88)90158-2. [DOI] [PubMed] [Google Scholar]
- Osumi T., Ishii N., Hijikata M., Kamijo K., Ozasa H., Furuta S., Miyazawa S., Kondo K., Inoue K., Kagamiyama H. Molecular cloning and nucleotide sequence of the cDNA for rat peroxisomal enoyl-CoA: hydratase-3-hydroxyacyl-CoA dehydrogenase bifunctional enzyme. J Biol Chem. 1985 Jul 25;260(15):8905–8910. [PubMed] [Google Scholar]
- Pramanik A., Pawar S., Antonian E., Schulz H. Five different enzymatic activities are associated with the multienzyme complex of fatty acid oxidation from Escherichia coli. J Bacteriol. 1979 Jan;137(1):469–473. doi: 10.1128/jb.137.1.469-473.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- STERN J. R., DEL CAMPILLO A., RAW I. Enzymes of fatty acid metabolism. I. General introduction; crystalline crotonase. J Biol Chem. 1956 Feb;218(2):971–983. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt S. K., Black P. N., Ragozzino M. M., Nunn W. D. Cloning, mapping, and expression of genes involved in the fatty acid-degradative multienzyme complex of Escherichia coli. J Bacteriol. 1984 May;158(2):535–542. doi: 10.1128/jb.158.2.535-542.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W. R., Thornton J. M. Recognition of super-secondary structure in proteins. J Mol Biol. 1984 Mar 15;173(4):487–512. [PubMed] [Google Scholar]
- Waterson R. M., Castellino F. J., Hass G. M., Hill R. L. Purification and characterization of crotonase from Clostridium acetobutylicum. J Biol Chem. 1972 Aug 25;247(16):5266–5271. [PubMed] [Google Scholar]
- Wierenga R. K., Hol W. G. Predicted nucleotide-binding properties of p21 protein and its cancer-associated variant. Nature. 1983 Apr 28;302(5911):842–844. doi: 10.1038/302842a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Youngleson Jonathan S., Santangelo Joseph D., Jones David T., Woods David R. Cloning and Expression of a Clostridium acetobutylicum Alcohol Dehydrogenase Gene in Escherichia coli. Appl Environ Microbiol. 1988 Mar;54(3):676–682. doi: 10.1128/aem.54.3.676-682.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappe H., Jones W. A., Jones D. T., Woods D. R. Structure of an endo-beta-1,4-glucanase gene from Clostridium acetobutylicum P262 showing homology with endoglucanase genes from Bacillus spp. Appl Environ Microbiol. 1988 May;54(5):1289–1292. doi: 10.1128/aem.54.5.1289-1292.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Duve C. Microbodies in the living cell. Sci Am. 1983 May;248(5):74–84. doi: 10.1038/scientificamerican0583-74. [DOI] [PubMed] [Google Scholar]


