Abstract
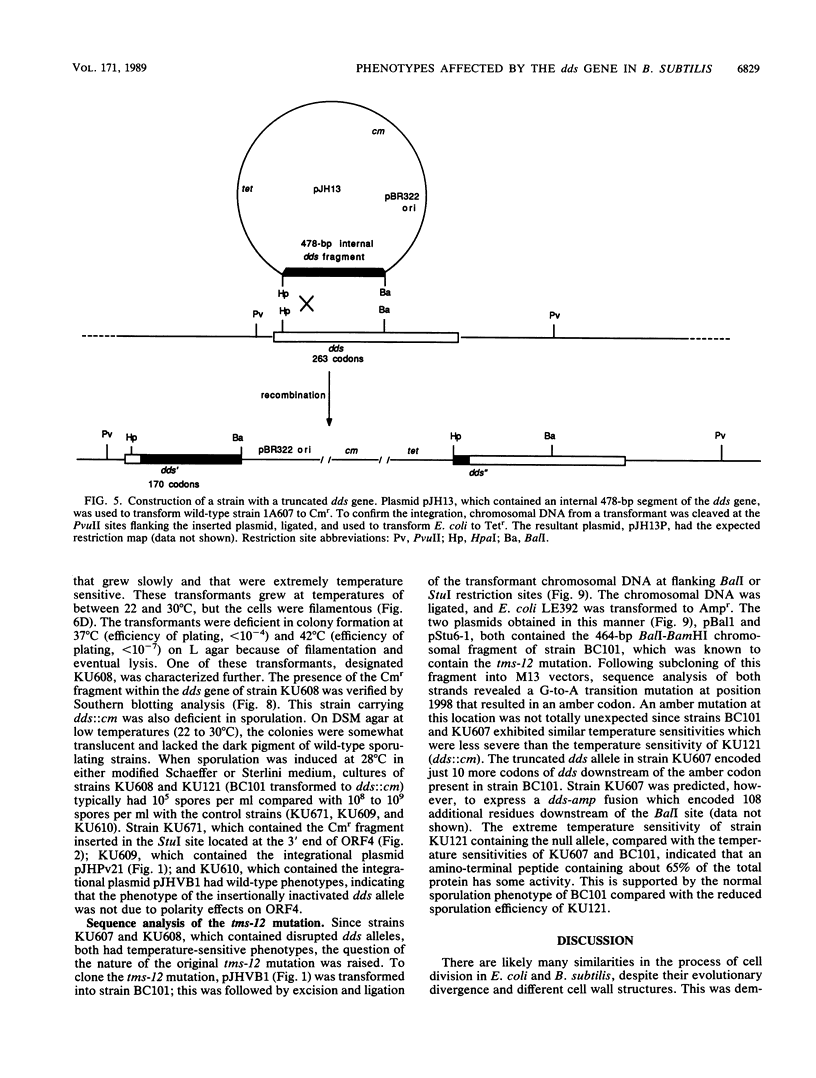
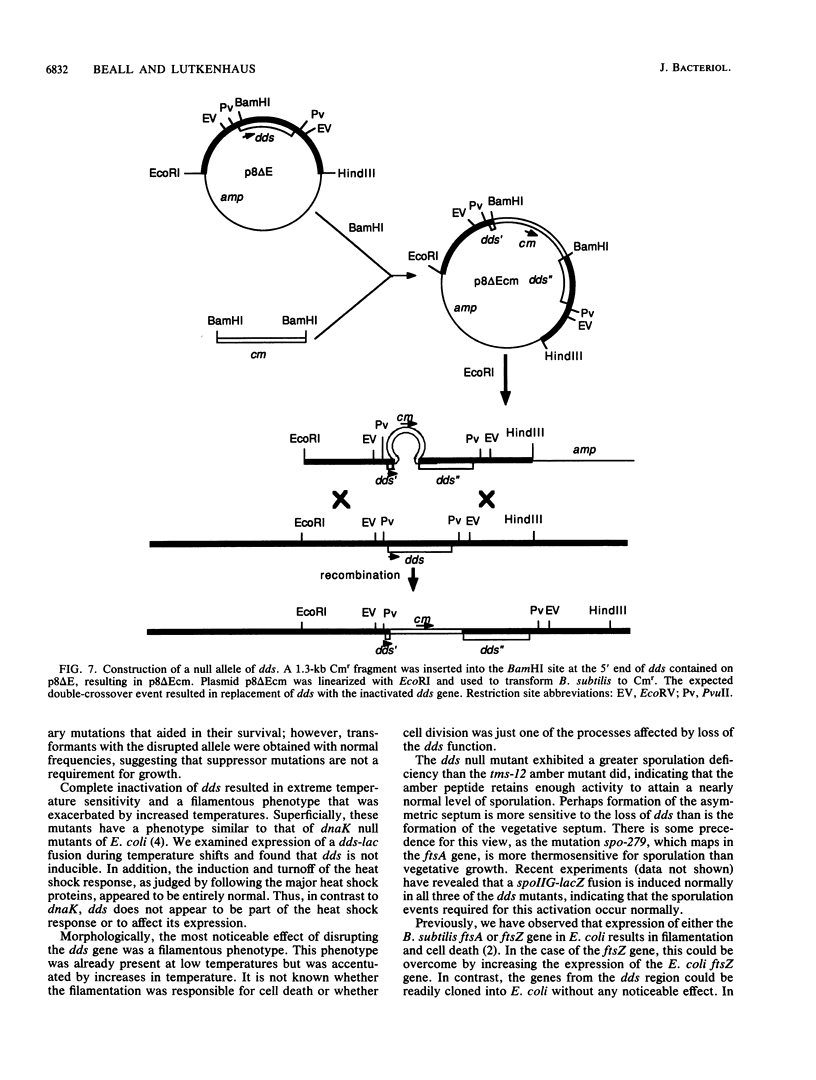
Located at 135 degrees on the Bacillus subtilis genetic map are several genes suspected to be involved in cell division and sporulation. Previously isolated mutations mapping at 135 degrees include the tms-12 mutation and mutations in the B. subtilis homologs of the Escherichia coli cell division genes ftsA and ftsZ. Previously, we cloned and sequenced the B. subtilis ftsA and ftsZ genes that are present on an 11-kilobase-pair EcoRI fragment and found that the gene products and organization of these two genes are conserved between the two bacterial species. We have since found that the mutation in the temperature-sensitive filamenting tms-12 mutant maps upstream of the ftsA gene on the same 11-kilobase-pair EcoRI fragment in a gene we designated dds. Sequence analysis of the dds gene and four other open reading frames upstream of ftsA revealed no significant homology to other known genes. It was found that the dds gene is not absolutely essential for viability since the dds gene could be insertionally inactivated. The dds null mutants grew slowly, were filamentous, and exhibited a reduced level of sporulation. Additionally, these mutants were extremely temperature sensitive and were unable to form colonies at 37 degrees C. Another insertion, which resulted in the elimination of 103 C-terminal residues, resulted in a temperature-sensitive phenotype less severe than that in the dds null mutant and similar to that in the known tms-12 mutant. The tms-12 mutation was cloned and sequenced, revealing a nonsense codon that was predicted to result in an amber fragment that was about 65% of the wild-type size (elimination of 93 C-terminal residues).
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler H. I., Fisher W. D., Cohen A., Hardigree A. A. MINIATURE escherichia coli CELLS DEFICIENT IN DNA. Proc Natl Acad Sci U S A. 1967 Feb;57(2):321–326. doi: 10.1073/pnas.57.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B., Lowe M., Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988 Oct;170(10):4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakefield X. O., Landman O. E. Temperature-sensitive divisionless mutant of Bacillus subtilis defective in the initiation of septation. J Bacteriol. 1973 Feb;113(2):985–998. doi: 10.1128/jb.113.2.985-998.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Walker G. C. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J Bacteriol. 1989 May;171(5):2337–2346. doi: 10.1128/jb.171.5.2337-2346.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister H., Wake R. G. Characterization and mapping of temperature-sensitive division initiation mutations of Bacillus subtilis. J Bacteriol. 1981 Feb;145(2):1042–1051. doi: 10.1128/jb.145.2.1042-1051.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland J. C., Marmur J. Identification of conserved genetic functions in Bacillus by use of temperature-sensitive mutants. Bacteriol Rev. 1968 Dec;32(4 Pt 1):302–312. [PMC free article] [PubMed] [Google Scholar]
- Corton J. C., Ward J. E., Jr, Lutkenhaus J. Analysis of cell division gene ftsZ (sulB) from gram-negative and gram-positive bacteria. J Bacteriol. 1987 Jan;169(1):1–7. doi: 10.1128/jb.169.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971 Mar 14;56(2):209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harry E. J., Wake R. G. Cloning and expression of a Bacillus subtilis division initiation gene for which a homolog has not been identified in another organism. J Bacteriol. 1989 Dec;171(12):6835–6839. doi: 10.1128/jb.171.12.6835-6839.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leighton T. J., Doi R. H. The stability of messenger ribonucleic acid during sporulation in Bacillus subtilis. J Biol Chem. 1971 May 25;246(10):3189–3195. [PubMed] [Google Scholar]
- Lutkenhaus J. Genetic analysis of bacterial cell division. Microbiol Sci. 1988 Mar;5(3):88–91. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nukushina J. I., Ikeda Y. Genetic analysis of the developmental processes during germination and outgrowth of Bacillus subtilis spores with temperature-sensitive mutants. Genetics. 1969 Sep;63(1):63–74. doi: 10.1093/genetics/63.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot P. J., Chak K. F., Bugaichuk U. D. Isolation and characterization of a clone of the spoVE locus of Bacillus subtilis. J Gen Microbiol. 1986 Jul;132(7):1875–1881. doi: 10.1099/00221287-132-7-1875. [DOI] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterlini J. M., Mandelstam J. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem J. 1969 Jun;113(1):29–37. doi: 10.1042/bj1130029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar M. A., Zahler S. A. Chromosomal insertions of Tn917 in Bacillus subtilis. J Bacteriol. 1986 Aug;167(2):530–534. doi: 10.1128/jb.167.2.530-534.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Young M. Genetic mapping of sporulation operons in Bacillus subtilis using a thermosensitive sporulation mutant. J Bacteriol. 1975 Jun;122(3):1109–1116. doi: 10.1128/jb.122.3.1109-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]






