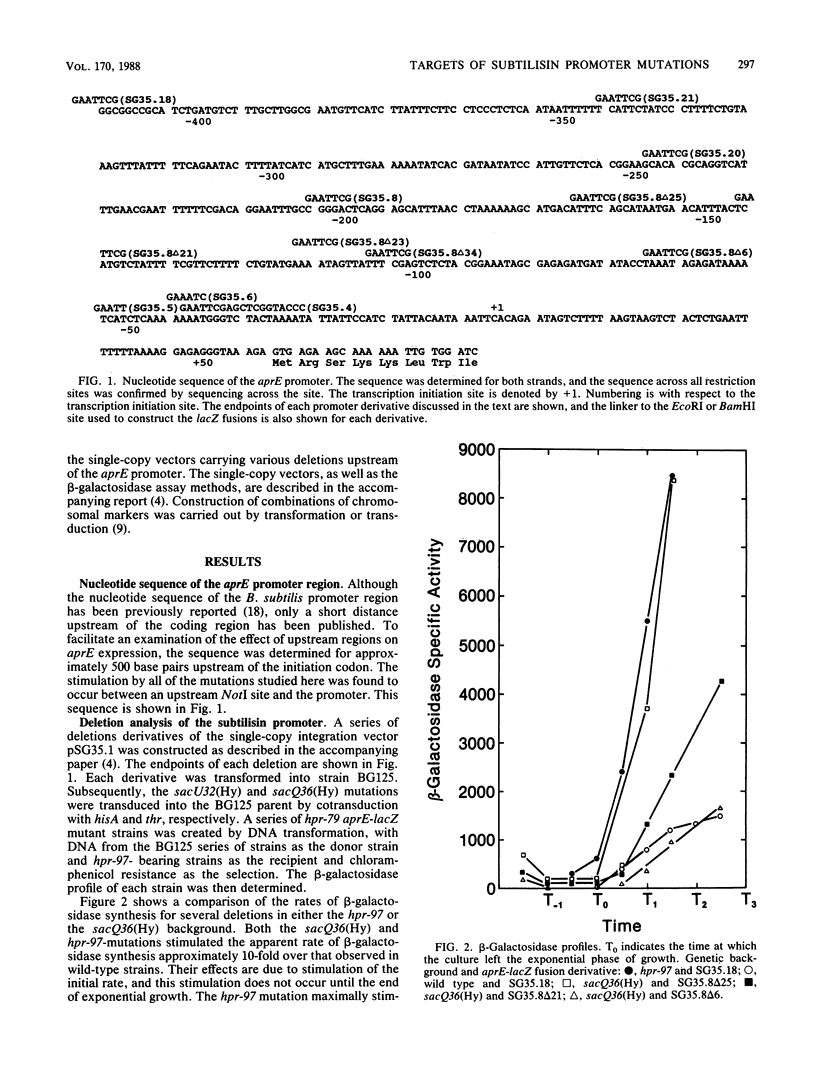
Abstract
A number of mutations have been described with pleiotropic effects on the expression of genes for degradative enzymes in Bacillus subtilis. The sacU32(Hy) and sacQ36(Hy) mutations increase the expression of a wide variety of enzymes that degrade biological polymers. The phenotypes caused by mutations at the hpr locus are more restricted; they are known to increase expression of the alkaline and neutral proteases. The alkaline protease (aprE) promoter was analyzed to determine the target site for stimulation by these loci. Deletion of upstream regions of the aprE promoter could abolish or greatly reduce stimulation by mutations at these loci. A region upstream of -200 was necessary for full stimulation by an hpr-97 mutation, whereas a region between -141 and -164 was necessary for full stimulation by the sacU32(Hy) and sacQ36(Hy) mutations. Northern analyses of mRNA preparations showed that the levels of aprE mRNA were increased in strains carrying the sacU32(Hy) or hpr-97 mutation. Moreover, primer extension analysis of these mRNA preparations revealed that the transcription start point was identical to that in a wild-type strain. We hypothesize that upstream activation of the subtilisin promoter mediated by these genes is a mechanism for global responses to a variety of nutritional conditions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amory A., Kunst F., Aubert E., Klier A., Rapoport G. Characterization of the sacQ genes from Bacillus licheniformis and Bacillus subtilis. J Bacteriol. 1987 Jan;169(1):324–333. doi: 10.1128/jb.169.1.324-333.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymerich S., Gonzy-Tréboul G., Steinmetz M. 5'-noncoding region sacR is the target of all identified regulation affecting the levansucrase gene in Bacillus subtilis. J Bacteriol. 1986 Jun;166(3):993–998. doi: 10.1128/jb.166.3.993-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambert R., Petit-Glatron M. F. Hyperproduction of exocellular levansucrase by Bacillus subtilis: examination of the phenotype of a sacUh strain. J Gen Microbiol. 1984 Dec;130(12):3143–3152. doi: 10.1099/00221287-130-12-3143. [DOI] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Perego M., Hoch J. A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988 Jan;170(1):289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur N. K., Dubnau E., Smith I. Characterization of a cloned Bacillus subtilis gene that inhibits sporulation in multiple copies. J Bacteriol. 1986 Nov;168(2):860–869. doi: 10.1128/jb.168.2.860-869.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gicquel-Sanzey B., Cossart P. Homologies between different procaryotic DNA-binding regulatory proteins and between their sites of action. EMBO J. 1982;1(5):591–595. doi: 10.1002/j.1460-2075.1982.tb01213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gussin G. N., Ronson C. W., Ausubel F. M. Regulation of nitrogen fixation genes. Annu Rev Genet. 1986;20:567–591. doi: 10.1146/annurev.ge.20.120186.003031. [DOI] [PubMed] [Google Scholar]
- Higerd T. B., Hoch J. A., Spizizen J. Hyperprotease-producing mutants of Bacillus subtilis. J Bacteriol. 1972 Nov;112(2):1026–1028. doi: 10.1128/jb.112.2.1026-1028.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A., Barat M., Anagnostopoulos C. Transformation and transduction in recombination-defective mutants of Bacillus subtilis. J Bacteriol. 1967 Jun;93(6):1925–1937. doi: 10.1128/jb.93.6.1925-1937.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst F., Pascal M., Lepesant-Kejzlarova J., Lepesant J. A., Billault A., Dedonder R. Pleiotropic mutations affecting sporulation conditions and the syntheses of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56(11-12):1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- Lepesant J. A., Kunst F., Lepesant-Kejzlarová J., Dedonder R. Chromosomal location of mutations affecting sucrose metabolism in Bacillus subtilis Marburg. Mol Gen Genet. 1972;118(2):135–160. doi: 10.1007/BF00267084. [DOI] [PubMed] [Google Scholar]
- Lepesant J. A., Lepesant-Kejzlarová J., Pascal M., Kunst F., Billault A., Dedonder R. Identification of the structural gene of levansucrase in Bacillus subtilis Marburg. Mol Gen Genet. 1974 Feb 6;128(3):213–221. doi: 10.1007/BF00267110. [DOI] [PubMed] [Google Scholar]
- Nagami Y., Tanaka T. Molecular cloning and nucleotide sequence of a DNA fragment from Bacillus natto that enhances production of extracellular proteases and levansucrase in Bacillus subtilis. J Bacteriol. 1986 Apr;166(1):20–28. doi: 10.1128/jb.166.1.20-28.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes. J Bacteriol. 1986 Oct;168(1):380–388. doi: 10.1128/jb.168.1.380-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. L., Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an In vitro-derived deletion mutation. J Bacteriol. 1984 May;158(2):411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kawata M., Nagami Y., Uchiyama H. prtR enhances the mRNA level of the Bacillus subtilis extracellular proteases. J Bacteriol. 1987 Jul;169(7):3044–3050. doi: 10.1128/jb.169.7.3044-3050.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Shimotsu H., Ferrari E., Henner D. J. Characterization and mapping of the Bacillus subtilis prtR gene. J Bacteriol. 1987 Jan;169(1):434–437. doi: 10.1128/jb.169.1.434-437.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]