Abstract
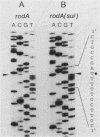
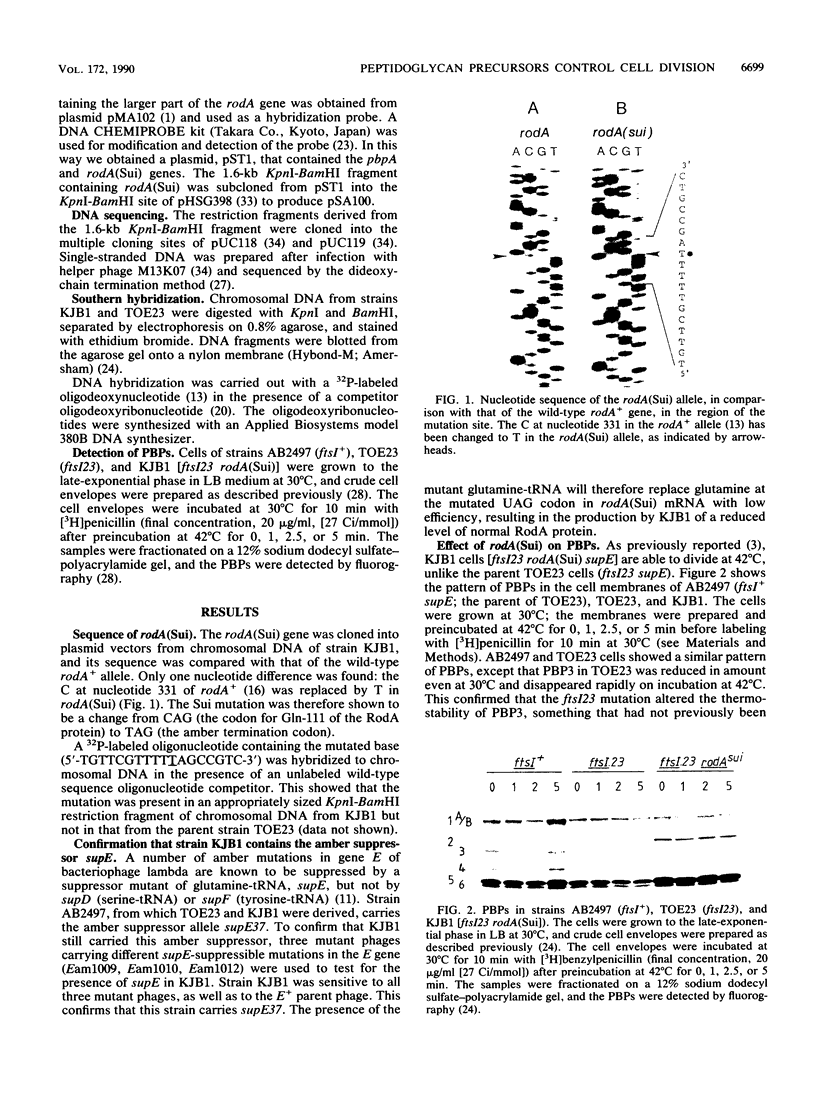
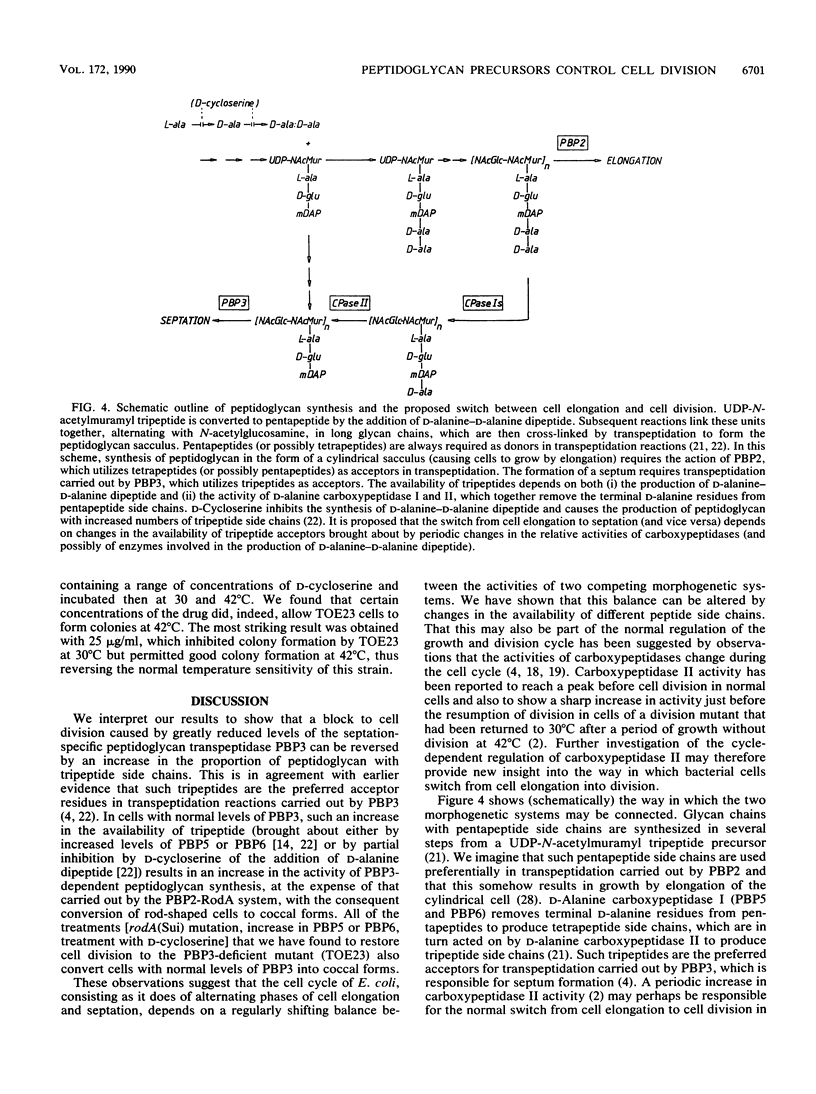
The rodA(Sui) mutation allows cell division to take place at 42 degrees C in ftsI23 mutant cells, which produce a thermolabile penicillin-binding protein 3 (PBP3, the septation-specific peptidoglycan transpeptidase). We show here that the mutation in rodA is a single-base change from a glutamine to a chain termination (amber) codon, and that an amber suppressor (supE) present in the strain restores the ability to produce a reduced level of normal RodA protein. The reduced level of RodA is accompanied by an increase in the levels of two other proteins (PBP2 and PBP5) encoded by genes in the rodA operon. We show that an increased level of PBP5 is by itself sufficient to restore cell division to ftsI23 cells at 42 degrees C. Two other treatments were found to restore division capacity to the mutant: an increase in PBP6 (which is a D-alanine carboxypeptidase like PBP5) or suitable concentrations of D-cycloserine. All of the above treatments have the effect of reducing the number of pentapeptide side chains in peptidoglycan and increasing the number of tripeptides. We conclude that the effect of the rodA(Sui) mutation is to indirectly increase the availability of tripeptide side chains, which are used preferentially by PBP3 as acceptors in transpeptidation. A change in the proportions of different kinds of peptide side chain in the peptidoglycan can therefore determine whether cells will divide.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Asoh S., Matsuzawa H., Matsuhashi M., Ohta T. Molecular cloning and characterization of the genes (pbpA and rodA) responsible for the rod shape of Escherichia coli K-12: analysis of gene expression with transposon Tn5 mutagenesis and protein synthesis directed by constructed plasmids. J Bacteriol. 1983 Apr;154(1):10–16. doi: 10.1128/jb.154.1.10-16.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B. D., Park J. T. Activity of three murein hydrolases during the cell division cycle of Escherichia coli K-12 as measured in toluene-treated cells. J Bacteriol. 1976 Jun;126(3):1250–1260. doi: 10.1128/jb.126.3.1250-1260.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Spratt B. G., Donachie W. D. Interaction between membrane proteins PBP3 and rodA is required for normal cell shape and division in Escherichia coli. J Bacteriol. 1986 Sep;167(3):1004–1008. doi: 10.1128/jb.167.3.1004-1008.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta G. A., Park J. T. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981 Jan;145(1):333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome-Smith J. K., Ioannidis I., Edelman A., Spratt B. G. Nucleotide sequences of the penicillin-binding protein 5 and 6 genes of Escherichia coli. Nucleic Acids Res. 1988 Feb 25;16(4):1617–1617. doi: 10.1093/nar/16.4.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broome-Smith J. K., Spratt B. G. Deletion of the penicillin-binding protein 6 gene of Escherichia coli. J Bacteriol. 1982 Nov;152(2):904–906. doi: 10.1128/jb.152.2.904-906.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie W. D., Begg K. Genes and the replication cycle of Escherichia coli. Res Microbiol. 1990 Jan;141(1):64–75. doi: 10.1016/0923-2508(90)90099-c. [DOI] [PubMed] [Google Scholar]
- Ferreira L. C., Keck W., Betzner A., Schwarz U. In vivo cell division gene product interactions in Escherichia coli K-12. J Bacteriol. 1987 Dec;169(12):5776–5781. doi: 10.1128/jb.169.12.5776-5781.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino F., Jung H. K., Ikeda M., Doi M., Wachi M., Matsuhashi M. New mutations fts-36, lts-33, and ftsW clustered in the mra region of the Escherichia coli chromosome induce thermosensitive cell growth and division. J Bacteriol. 1989 Oct;171(10):5523–5530. doi: 10.1128/jb.171.10.5523-5530.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura I. Structure and inherent properties of the bacteriophage lambda head shell. V. Amber mutants in gene E. J Mol Biol. 1986 Aug 20;190(4):577–586. doi: 10.1016/0022-2836(86)90243-3. [DOI] [PubMed] [Google Scholar]
- Markiewicz Z., Broome-Smith J. K., Schwarz U., Spratt B. G. Spherical E. coli due to elevated levels of D-alanine carboxypeptidase. Nature. 1982 Jun 24;297(5868):702–704. doi: 10.1038/297702a0. [DOI] [PubMed] [Google Scholar]
- Matsuhashi M., Wachi M., Ishino F. Machinery for cell growth and division: penicillin-binding proteins and other proteins. Res Microbiol. 1990 Jan;141(1):89–103. doi: 10.1016/0923-2508(90)90101-u. [DOI] [PubMed] [Google Scholar]
- Matsuzawa H., Asoh S., Kunai K., Muraiso K., Takasuga A., Ohta T. Nucleotide sequence of the rodA gene, responsible for the rod shape of Escherichia coli: rodA and the pbpA gene, encoding penicillin-binding protein 2, constitute the rodA operon. J Bacteriol. 1989 Jan;171(1):558–560. doi: 10.1128/jb.171.1.558-560.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mirelman D., Yashouv-Gan Y., Nuchamovitz Y., Rozenhak S., Ron E. Z. Murein biosynthesis during a synchromous cell cycle of Escherichia coli B. J Bacteriol. 1978 May;134(2):458–461. doi: 10.1128/jb.134.2.458-461.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirelman D., Yashouv-Gan Y., Schwarz U. Regulation of murein biosynthesis and septum formation in filamentous cells of Escherichia coli PAT 84. J Bacteriol. 1977 Mar;129(3):1593–1600. doi: 10.1128/jb.129.3.1593-1600.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozari G., Rahbar S., Wallace R. B. Discrimination among the transcripts of the allelic human beta-globin genes beta A, beta S and beta C using oligodeoxynucleotide hybridization probes. Gene. 1986;43(1-2):23–28. doi: 10.1016/0378-1119(86)90004-1. [DOI] [PubMed] [Google Scholar]
- Pisabarro A. G., Prats R., Váquez D., Rodríguez-Tébar A. Activity of penicillin-binding protein 3 from Escherichia coli. J Bacteriol. 1986 Oct;168(1):199–206. doi: 10.1128/jb.168.1.199-206.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poverenny A. M., Podgorodnichenko V. K., Bryksina L. E., Monastyrskaya G. S., Sverdlov E. D. Immunochemical approaches to DNA structure investigation--I. Immunochemical identification of the product of cytosine modification with bisulphite and O-methylhydroxylamine mixture. Mol Immunol. 1979 May;16(5):313–316. doi: 10.1016/0161-5890(79)90132-9. [DOI] [PubMed] [Google Scholar]
- Reed K. C., Mann D. A. Rapid transfer of DNA from agarose gels to nylon membranes. Nucleic Acids Res. 1985 Oct 25;13(20):7207–7221. doi: 10.1093/nar/13.20.7207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson A. C., Begg K. J., Sweeney J., Condie A., Donachie W. D. Mapping and characterization of mutants of the Escherichia coli cell division gene, ftsA. Mol Microbiol. 1988 Sep;2(5):581–588. doi: 10.1111/j.1365-2958.1988.tb00066.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Biochemical and genetical approaches to the mechanism of action of penicillin. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):273–283. doi: 10.1098/rstb.1980.0045. [DOI] [PubMed] [Google Scholar]
- Spratt B. G., Boyd A., Stoker N. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from the lip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J Bacteriol. 1980 Aug;143(2):569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt B. G. Properties of the penicillin-binding proteins of Escherichia coli K12,. Eur J Biochem. 1977 Jan;72(2):341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- Stoker N. G., Pratt J. M., Spratt B. G. Identification of the rodA gene product of Escherichia coli. J Bacteriol. 1983 Aug;155(2):854–859. doi: 10.1128/jb.155.2.854-859.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase I., Ishino F., Wachi M., Kamata H., Doi M., Asoh S., Matsuzawa H., Ohta T., Matsuhashi M. Genes encoding two lipoproteins in the leuS-dacA region of the Escherichia coli chromosome. J Bacteriol. 1987 Dec;169(12):5692–5699. doi: 10.1128/jb.169.12.5692-5699.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wachi M., Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989 Jun;171(6):3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]