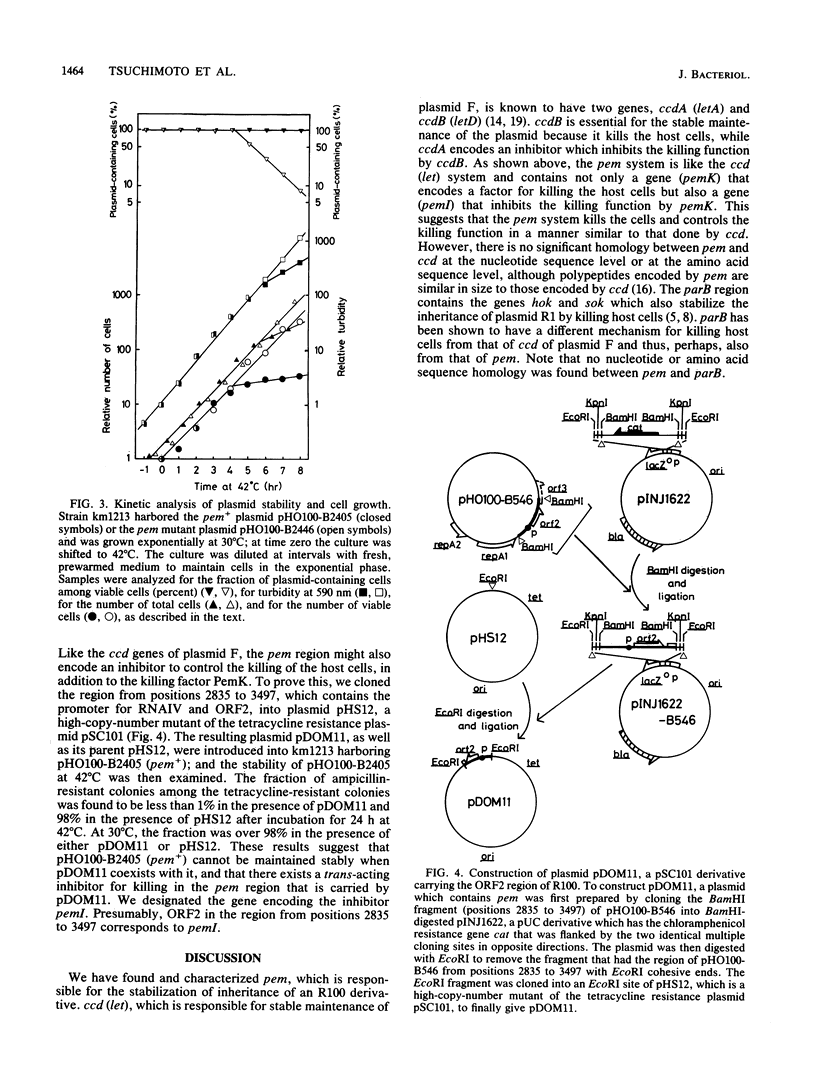
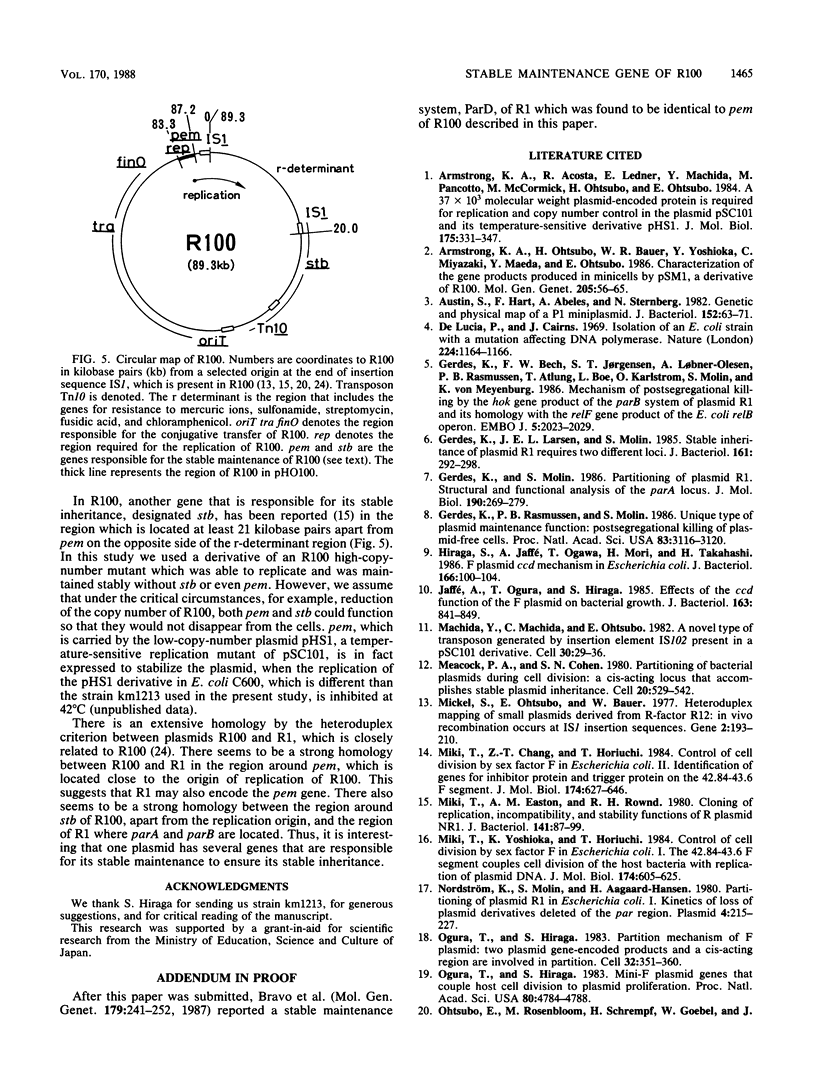
Abstract
Plasmid R100 was found to have two genes, designated pemK and pemI, that were responsible for its stable inheritance during cell division. They are located near the region that is essential for autonomous replication. Under conditions that inhibit replication of R100 derivatives, the plasmid containing these pem genes gave only a few segregants in viable cells and increased the number of nonviable cells in the population, suggesting that a product from the pem region stabilized the plasmid by killing plasmid-free segregants. Inactivation of one of the two translational open reading frames in the pem region caused the loss of the killing function, and thus, the open reading frame is a gene designated pemK, which encodes the killing factor. The coexistence of the pem+ plasmid with a high-copy-number plasmid carrying the other open reading frame inhibited stabilization, and thus, the second open reading frame is a gene designated pemI, which encodes the inhibitor which might control the killing function of pemK. It is likely that the two open reading frames were transcribed from a promoter. There were no significant homologies in DNA sequences between the pem gene of R100 and the genes previously shown to be responsible for the stable inheritance of the other plasmids.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K. A., Acosta R., Ledner E., Machida Y., Pancotto M., McCormick M., Ohtsubo H., Ohtsubo E. A 37 X 10(3) molecular weight plasmid-encoded protein is required for replication and copy number control in the plasmid pSC101 and its temperature-sensitive derivative pHS1. J Mol Biol. 1984 May 25;175(3):331–348. doi: 10.1016/0022-2836(84)90352-8. [DOI] [PubMed] [Google Scholar]
- Armstrong K. A., Ohtsubo H., Bauer W. R., Yoshioka Y., Miyazaki C., Maeda Y., Ohtsubo E. Characterization of the gene products produced in minicells by pSM1, a derivative of R100. Mol Gen Genet. 1986 Oct;205(1):56–65. doi: 10.1007/BF02428032. [DOI] [PubMed] [Google Scholar]
- Austin S., Hart F., Abeles A., Sternberg N. Genetic and physical map of a P1 miniplasmid. J Bacteriol. 1982 Oct;152(1):63–71. doi: 10.1128/jb.152.1.63-71.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lucia P., Cairns J. Isolation of an E. coli strain with a mutation affecting DNA polymerase. Nature. 1969 Dec 20;224(5225):1164–1166. doi: 10.1038/2241164a0. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Bech F. W., Jørgensen S. T., Løbner-Olesen A., Rasmussen P. B., Atlung T., Boe L., Karlstrom O., Molin S., von Meyenburg K. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 1986 Aug;5(8):2023–2029. doi: 10.1002/j.1460-2075.1986.tb04459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Larsen J. E., Molin S. Stable inheritance of plasmid R1 requires two different loci. J Bacteriol. 1985 Jan;161(1):292–298. doi: 10.1128/jb.161.1.292-298.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K., Molin S. Partitioning of plasmid R1. Structural and functional analysis of the parA locus. J Mol Biol. 1986 Aug 5;190(3):269–279. doi: 10.1016/0022-2836(86)90001-x. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraga S., Jaffé A., Ogura T., Mori H., Takahashi H. F plasmid ccd mechanism in Escherichia coli. J Bacteriol. 1986 Apr;166(1):100–104. doi: 10.1128/jb.166.1.100-104.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffé A., Ogura T., Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985 Sep;163(3):841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida Y., Machida C., Ohtsubo E. A novel type of transposon generated by insertion element IS102 present in a pSC101 derivative. Cell. 1982 Aug;30(1):29–36. doi: 10.1016/0092-8674(82)90008-3. [DOI] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Miki T., Chang Z. T., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. II. Identification of genes for inhibitor protein and trigger protein on the 42.84-43.6 F segment. J Mol Biol. 1984 Apr 25;174(4):627–646. doi: 10.1016/0022-2836(84)90087-1. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T., Yoshioka K., Horiuchi T. Control of cell division by sex factor F in Escherichia coli. I. The 42.84-43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. J Mol Biol. 1984 Apr 25;174(4):605–625. doi: 10.1016/0022-2836(84)90086-x. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid. 1980 Sep;4(2):215–227. doi: 10.1016/0147-619x(80)90011-6. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Ohtsubo E., Rosenbloom M., Schrempf H., Goebel W., Rosen J. Site specific recombination involved in the generation of small plasmids. Mol Gen Genet. 1978 Feb 16;159(2):131–141. doi: 10.1007/BF00270886. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ryder T. B., Maeda Y., Armstrong K., Ohtsubo E. DNA replication of the resistance plasmid R100 and its control. Adv Biophys. 1986;21:115–133. doi: 10.1016/0065-227x(86)90018-3. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ohtsubo H., Ohtsubo E. The nucleotide sequence of the region surrounding the replication origin of an R100 resistance factor derivative. Mol Gen Genet. 1979 Mar 27;171(3):287–293. doi: 10.1007/BF00267583. [DOI] [PubMed] [Google Scholar]
- Rosen J., Ryder T., Ohtsubo H., Ohtsubo E. Role of RNA transcripts in replication incompatibility and copy number control in antibiotic resistance plasmid derivatives. Nature. 1981 Apr 30;290(5809):794–797. doi: 10.1038/290794a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Tanimoto K., Iino T., Ohtsubo H., Ohtsubo E. Identification of a gene, tir of R100, functionally homologous to the F3 gene of F in the inhibition of RP4 transfer. Mol Gen Genet. 1985;198(2):356–357. doi: 10.1007/BF00383019. [DOI] [PubMed] [Google Scholar]
- Tsutsui H., Matsubara K. Replication control and switch-off function as observed with a mini-F factor plasmid. J Bacteriol. 1981 Aug;147(2):509–516. doi: 10.1128/jb.147.2.509-516.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka Y., Ohtsubo H., Ohtsubo E. Repressor gene finO in plasmids R100 and F: constitutive transfer of plasmid F is caused by insertion of IS3 into F finO. J Bacteriol. 1987 Feb;169(2):619–623. doi: 10.1128/jb.169.2.619-623.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


