Abstract
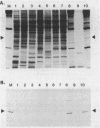
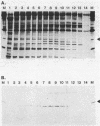
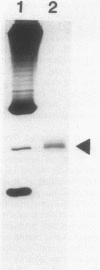
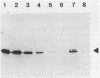
RNA polymerase preparations isolated from vegetatively growing Bacillus subtilis cells contain the core subunits beta, beta', and alpha, together with multiple sigma factors and other core-associated polypeptides such as delta, omega 1, and omega 2. We have developed an improved, large-scale purification procedure that yields RNA polymerase fractions enriched in both the sigma 28 and delta proteins. These fractions have been used to isolate sigma 28 protein for biochemical characterization and for preparation of highly specific anti-sigma 28 antisera. The amino acid composition of purified sigma 28 protein and the amino acid sequences of tryptic peptide fragments have been determined. Anti-sigma 28 antisera specifically inhibit transcription by the purified sigma 28 -dependent RNA polymerase, yet do not affect transcription by sigma 43 -dependent RNA polymerase. Immunochemical analysis confirms that the sigma 28 protein copurifies with total RNA polymerase activity through the majority of the purification procedure and allows the steps when sigma 28 protein is lost to be identified and optimized. Immunochemical techniques have also been used to monitor the structure and abundance of the sigma 28 protein in vivo. A single form of antibody-reactive protein was detected by two-dimensional gel electrophoresis-isoelectric focusing. Its abundance corresponds to a maximal content of 220 molecules of sigma 28 per B. subtilis cell during late-logarithmic-phase growth.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amemiya K., Wu C. W., Shapiro L. Caulobacter crescentus RNA polymerase. Purification and characterization of holoenzyme and core polymerase. J Biol Chem. 1977 Jun 25;252(12):4157–4165. [PubMed] [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Binnie C., Lampe M., Losick R. Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Toxicity of an overproduced foreign gene product in Escherichia coli and its use in plasmid vectors for the selection of transcription terminators. Gene. 1984 Feb;27(2):161–172. doi: 10.1016/0378-1119(84)90137-9. [DOI] [PubMed] [Google Scholar]
- Burgess R. R., Travers A. A., Dunn J. J., Bautz E. K. Factor stimulating transcription by RNA polymerase. Nature. 1969 Jan 4;221(5175):43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- Carter H. L., 3rd, Moran C. P., Jr New RNA polymerase sigma factor under spo0 control in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9438–9442. doi: 10.1073/pnas.83.24.9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin M. J., Nierman W. C., Wiggs J., Neff N. A quantitative assay for bacterial RNA polymerases. J Biol Chem. 1979 Oct 25;254(20):10061–10069. [PubMed] [Google Scholar]
- Davison B. L., Leighton T., Rabinowitz J. C. Purification of Bacillus subtilis RNA polymerase with heparin-agarose. In vitro transcription of phi 29 DNA. J Biol Chem. 1979 Sep 25;254(18):9220–9226. [PubMed] [Google Scholar]
- Duffy J. J., Geiduschek E. P. RNA polymerase from phage SP01-infected and uninfected Bacillus subtilis. J Biol Chem. 1975 Jun 25;250(12):4530–4541. [PubMed] [Google Scholar]
- Duncan M. L., Kalman S. S., Thomas S. M., Price C. W. Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilis RNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function. J Bacteriol. 1987 Feb;169(2):771–778. doi: 10.1128/jb.169.2.771-778.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Wiggs J. L., Chamberlin M. J. Nucleotide sequences of two Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase. Nucleic Acids Res. 1981 Nov 25;9(22):5991–6000. doi: 10.1093/nar/9.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman A. D., Erickson J. W., Gross C. A. The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell. 1984 Sep;38(2):383–390. doi: 10.1016/0092-8674(84)90493-8. [DOI] [PubMed] [Google Scholar]
- Hager D. A., Burgess R. R. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal Biochem. 1980 Nov 15;109(1):76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Lang N., Losick R. A sporulation-induced sigma-like regulatory protein from B. subtilis. Cell. 1981 Feb;23(2):615–624. doi: 10.1016/0092-8674(81)90157-4. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature. 1979 Nov 15;282(5736):256–260. doi: 10.1038/282256a0. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7000–7004. doi: 10.1073/pnas.77.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrikson R. L., Krueger E. T., Keim P. S. Amino acid sequence of phospholipase A2-alpha from the venom of Crotalus adamanteus. A new classification of phospholipases A2 based upon structural determinants. J Biol Chem. 1977 Jul 25;252(14):4913–4921. [PubMed] [Google Scholar]
- Helmann J. D., Márquez L. M., Chamberlin M. J. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J Bacteriol. 1988 Apr;170(4):1568–1574. doi: 10.1128/jb.170.4.1568-1574.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Hirschman J., Wong P. K., Sei K., Keener J., Kustu S. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc Natl Acad Sci U S A. 1985 Nov;82(22):7525–7529. doi: 10.1073/pnas.82.22.7525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Hunt T. P., Magasanik B. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8453–8457. doi: 10.1073/pnas.82.24.8453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M., Lampe M., Ray C., Schafer W., Moran C. P., Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987 Aug;169(8):3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis A. S. Single hydrolysis method for all amino acids, including cysteine and tryptophan. Methods Enzymol. 1983;91:26–36. doi: 10.1016/s0076-6879(83)91007-8. [DOI] [PubMed] [Google Scholar]
- Jaehning J. A., Wiggs J. L., Chamberlin M. J. Altered promoter selection by a novel form of Bacillus subtilis RNA polymerase. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5470–5474. doi: 10.1073/pnas.76.11.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. C., DeBacker M., Boezi J. A. Deoxyribonucleic acid-dependent ribonucleic acid polymerase of Pseudomonas putida. J Biol Chem. 1971 Mar 10;246(5):1222–1232. [PubMed] [Google Scholar]
- Johnson W. C., Moran C. P., Jr, Losick R. Two RNA polymerase sigma factors from Bacillus subtilis discriminate between overlapping promoters for a developmentally regulated gene. Nature. 1983 Apr 28;302(5911):800–804. doi: 10.1038/302800a0. [DOI] [PubMed] [Google Scholar]
- Kane C. M., Linn S. Purification and characterization of an apurinic/apyrimidinic endonuclease from HeLa cells. J Biol Chem. 1981 Apr 10;256(7):3405–3414. [PubMed] [Google Scholar]
- Kenney T. J., Moran C. P., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987 Jul;169(7):3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knopf U. C. The nucleoside triphosphate-ribonucleic acid nucleotidyltransferase (EC 2.7.7.6) of Agrobacterium tumefaciens (Smith and Townsend) Conn. Purification and properties of the enzyme from the tumorigenic strain B6806. Biochem J. 1974 Dec;143(3):511–520. doi: 10.1042/bj1430511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konigsberg W. H., Henderson L. Removal of sodium dodecyl sulfate from proteins by ion-pair extraction. Methods Enzymol. 1983;91:254–259. doi: 10.1016/s0076-6879(83)91022-4. [DOI] [PubMed] [Google Scholar]
- LaBell T. L., Trempy J. E., Haldenwang W. G. Sporulation-specific sigma factor sigma 29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci U S A. 1987 Apr;84(7):1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill U. I., Kniep-Behrendt E. M., Bock L., Hartmann G. R. Purification and characterization of the DNA-dependent RNA polymerase and its subunit sigma from Micrococcus luteus. Hoppe Seylers Z Physiol Chem. 1977 Dec;358(12):1591–1603. doi: 10.1515/bchm2.1977.358.2.1591. [DOI] [PubMed] [Google Scholar]
- Losick R., Youngman P., Piggot P. J. Genetics of endospore formation in Bacillus subtilis. Annu Rev Genet. 1986;20:625–669. doi: 10.1146/annurev.ge.20.120186.003205. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- O'Farrell P. H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975 May 25;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- Stein S., Brink L. Amino acid analysis of proteins and peptides at the picomole level: the fluorescamine amino acid analyzer. Methods Enzymol. 1981;79(Pt B):20–25. doi: 10.1016/s0076-6879(81)79008-6. [DOI] [PubMed] [Google Scholar]
- Tijan R., Pero J. Bacteriophage SP01 regulatory proteins directing late gene transcription in vitro. Nature. 1976 Aug 26;262(5571):753–757. doi: 10.1038/262753a0. [DOI] [PubMed] [Google Scholar]
- Trempy J. E., Bonamy C., Szulmajster J., Haldenwang W. G. Bacillus subtilis sigma factor sigma 29 is the product of the sporulation-essential gene spoIIG. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4189–4192. doi: 10.1073/pnas.82.12.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiggs J. L., Gilman M. Z., Chamberlin M. J. Heterogeneity of RNA polymerase in Bacillus subtilis: evidence for an additional sigma factor in vegetative cells. Proc Natl Acad Sci U S A. 1981 May;78(5):2762–2766. doi: 10.1073/pnas.78.5.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkin M. D. Structure and function in a Bacillus subtilis sporulation-specific sigma factor: molecular nature of mutations in spoIIAC. J Gen Microbiol. 1987 Mar;133(3):475–481. doi: 10.1099/00221287-133-3-475. [DOI] [PubMed] [Google Scholar]
- Zillig W., Zechel K., Halbwachs H. J. A new method of large scale preparation of highly purified DNA-dependent RNA-polymerase from E. coli. Hoppe Seylers Z Physiol Chem. 1970 Feb;351(2):221–224. doi: 10.1515/bchm2.1970.351.1.221. [DOI] [PubMed] [Google Scholar]