Abstract
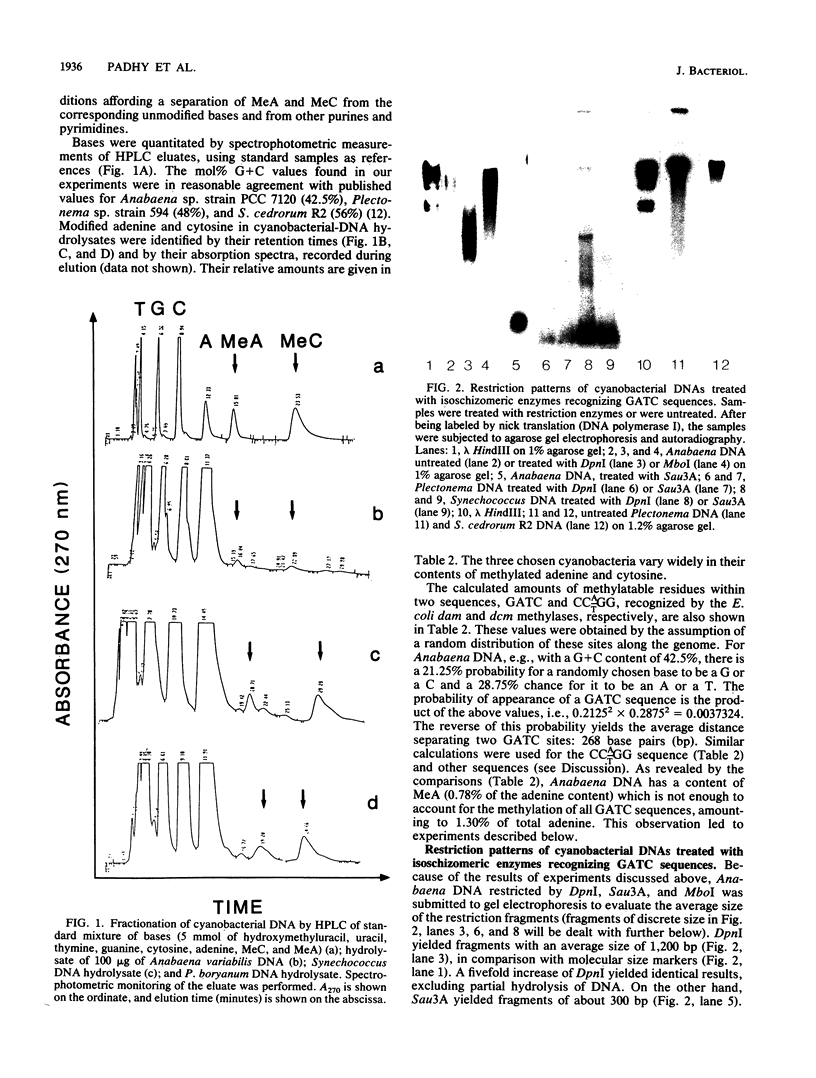
The DNAs of strains of three cyanobacterial genera (Anabaena, Plectonema, and Synechococcus) were found to be partially or fully resistant to many restriction endonucleases. This could be due to the absence of specific sequences or to modifications, rendering given sequences resistant to cleavage. The latter explanation is substantiated by the content of N6-methyladenine and 5-methylcytosine in these genomes, which is high in comparison with that in other bacterial genomes. dcm- and dam-like methylases are present in the three strains (based on the restriction patterns obtained with the appropriate isoschizomeric enzymes). Their contribution to the overall content of methyladenine and methylcytosine in the genomes was calculated. Partial methylation of GATC sequences was observed in Anabaena DNA. In addition, the GATC methylation patterns might not have been random in the three cyanobacterial DNA preparations, as revealed by the appearance of discrete fragments (possibly of plasmid origin) withstanding cleavage by DpnI (which requires the presence of methyladenine in the GATC sequence).
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barbeyron T., Kean K., Forterre P. DNA adenine methylation of GATC sequences appeared recently in the Escherichia coli lineage. J Bacteriol. 1984 Nov;160(2):586–590. doi: 10.1128/jb.160.2.586-590.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Blumenthal R. M., Gingeras T. R. The isolation and characterization of the Escherichia coli DNA adenine methylase (dam) gene. Nucleic Acids Res. 1983 Feb 11;11(3):837–851. doi: 10.1093/nar/11.3.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks J. E., Roberts R. J. Modification profiles of bacterial genomes. Nucleic Acids Res. 1982 Feb 11;10(3):913–934. doi: 10.1093/nar/10.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzby J. S., Porter R. D., Stevens S. E., Jr Plasmid transformation in Agmenellum quadruplicatum PR-6: construction of biphasic plasmids and characterization of their transformation properties. J Bacteriol. 1983 Jun;154(3):1446–1450. doi: 10.1128/jb.154.3.1446-1450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coene M., Cocito C. A microanalytical procedure for determination of the base composition of DNA. Eur J Biochem. 1985 Aug 1;150(3):475–479. doi: 10.1111/j.1432-1033.1985.tb09046.x. [DOI] [PubMed] [Google Scholar]
- Eick D., Fritz H. J., Doerfler W. Quantitative determination of 5-methylcytosine in DNA by reverse-phase high-performance liquid chromatography. Anal Biochem. 1983 Nov;135(1):165–171. doi: 10.1016/0003-2697(83)90746-7. [DOI] [PubMed] [Google Scholar]
- Golden G. M., Guzek D. B., Harris R. R., McKie J. E., Potts R. O. Lipid thermotropic transitions in human stratum corneum. J Invest Dermatol. 1986 Mar;86(3):255–259. doi: 10.1111/1523-1747.ep12285373. [DOI] [PubMed] [Google Scholar]
- Hattman S., Wilkinson J., Swinton D., Schlagman S., Macdonald P. M., Mosig G. Common evolutionary origin of the phage T4 dam and host Escherichia coli dam DNA-adenine methyltransferase genes. J Bacteriol. 1985 Nov;164(2):932–937. doi: 10.1128/jb.164.2.932-937.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattman S., van Ormondt H., de Waard A. Sequence specificity of the wild-type dam+) and mutant (damh) forms of bacteriophage T2 DNA adenine methylase. J Mol Biol. 1978 Mar 5;119(3):361–376. doi: 10.1016/0022-2836(78)90219-x. [DOI] [PubMed] [Google Scholar]
- Herrero A., Elhai J., Hohn B., Wolk C. P. Infrequent cleavage of cloned Anabaena variabilis DNA by restriction endonucleases from A. variabilis. J Bacteriol. 1984 Nov;160(2):781–784. doi: 10.1128/jb.160.2.781-784.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero A., Wolk C. P. Genetic mapping of the chromosome of the cyanobacterium, Anabaena variabilis. Proximity of the structural genes for nitrogenase and ribulose-bisphosphate carboxylase. J Biol Chem. 1986 Jun 15;261(17):7748–7754. [PubMed] [Google Scholar]
- Karreman C., Tandeau de Marsac N., de Waard A. Isolation of a deoxycytidylate methyl transferase capable of protecting DNA uniquely against cleavage by endonuclease R.Aqu I (isoschizomer of Ava I). Nucleic Acids Res. 1986 Jul 11;14(13):5199–5205. doi: 10.1093/nar/14.13.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler C., Neumaier P. S., Wolf W. Recognition sequences of restriction endonucleases and methylases--a review. Gene. 1985;33(1):1–102. doi: 10.1016/0378-1119(85)90119-2. [DOI] [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Complementary specificity of restriction endonucleases of Diplococcus pneumoniae with respect to DNA methylation. J Mol Biol. 1977 Jul;114(1):153–168. doi: 10.1016/0022-2836(77)90289-3. [DOI] [PubMed] [Google Scholar]
- Lambert G. R., Carr N. G. Resistance of DNA from filamentous and unicellular cyanobacteria to restriction endonuclease cleavage. Biochim Biophys Acta. 1984 Feb 24;781(1-2):45–55. doi: 10.1016/0167-4781(84)90122-2. [DOI] [PubMed] [Google Scholar]
- Lau R. H., Doolittle W. F. Covalently closed circular DNAs in closely related unicellular cyanobacteria. J Bacteriol. 1979 Jan;137(1):648–652. doi: 10.1128/jb.137.1.648-652.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau R. H., Sapienza C., Doolittle W. F. Cyanobacterial plasmids: their widespread occurrence, and the existence of regions of homology between plasmids in the same and different species. Mol Gen Genet. 1980 Apr;178(1):203–211. doi: 10.1007/BF00267230. [DOI] [PubMed] [Google Scholar]
- Mannarelli B. M., Balganesh T. S., Greenberg B., Springhorn S. S., Lacks S. A. Nucleotide sequence of the Dpn II DNA methylase gene of Streptococcus pneumoniae and its relationship to the dam gene of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4468–4472. doi: 10.1073/pnas.82.13.4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Urieli S., Pollack Y., Gruenbaum Y., Glaser G. Studies on the biological role of dna methylation; IV. Mode of methylation of DNA in E. coli cells. Nucleic Acids Res. 1980 Apr 25;8(8):1783–1792. doi: 10.1093/nar/8.8.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaston J., Duyvesteyn G. C., de Waard A. Nostoc PCC7524, a cyanobacterium which contains five sequence-specific deoxyribonucleases. Gene. 1982 Nov;20(1):103–110. doi: 10.1016/0378-1119(82)90091-9. [DOI] [PubMed] [Google Scholar]
- Reaston J., van den Hondel C. A., van Arkel G. A., Stewart W. D. A physical map of plasmid pDU1 from the cyanobacterium Nostoc PCC 7524. Plasmid. 1982 Jan;7(1):101–104. doi: 10.1016/0147-619x(82)90032-4. [DOI] [PubMed] [Google Scholar]
- Roberts T. M., Koths K. E. The blue-green alga agmenellum quadruplicatum contains covalently closed DNA circles. Cell. 1976 Dec;9(4 Pt 1):551–557. doi: 10.1016/0092-8674(76)90037-4. [DOI] [PubMed] [Google Scholar]
- Simon R. D. Survey of extrachromosomal DNA found in the filamentous cyanobacteria. J Bacteriol. 1978 Oct;136(1):414–418. doi: 10.1128/jb.136.1.414-418.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanier R. Y., Cohen-Bazire G. Phototrophic prokaryotes: the cyanobacteria. Annu Rev Microbiol. 1977;31:225–274. doi: 10.1146/annurev.mi.31.100177.001301. [DOI] [PubMed] [Google Scholar]
- Sternberg N. Evidence that adenine methylation influences DNA-protein interactions in Escherichia coli. J Bacteriol. 1985 Oct;164(1):490–493. doi: 10.1128/jb.164.1.490-493.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyf M., Avraham-Haetzni K., Reifman A., Shlomai J., Kaplan F., Oppenheim A., Razin A. DNA methylation pattern is determined by the intracellular level of the methylase. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3278–3282. doi: 10.1073/pnas.81.11.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urieli-Shoval S., Gruenbaum Y., Razin A. Sequence and substrate specificity of isolated DNA methylases from Escherichia coli C. J Bacteriol. 1983 Jan;153(1):274–280. doi: 10.1128/jb.153.1.274-280.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Belozersky A. N., Kokurina N. A., Kadirova D. X. 5-methylcytosine and 6-methylamino-purine in bacterial DNA. Nature. 1968 Jun 15;218(5146):1066–1067. doi: 10.1038/2181066a0. [DOI] [PubMed] [Google Scholar]
- Whitehead P. R., Brown N. L. AhaIII: a restriction endonuclease with a recognition sequence containing only A:T basepairs. FEBS Lett. 1982 Jul 5;143(2):296–300. doi: 10.1016/0014-5793(82)80120-8. [DOI] [PubMed] [Google Scholar]
- Wolk C. P., Vonshak A., Kehoe P., Elhai J. Construction of shuttle vectors capable of conjugative transfer from Escherichia coli to nitrogen-fixing filamentous cyanobacteria. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1561–1565. doi: 10.1073/pnas.81.5.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Waard A., van Beveren C. P., Duyvesteyn M., van Ormondt H. Two sequence-specific endonucleases from Anabaena oscillariodes. FEBS Lett. 1979 May 1;101(1):71–76. [PubMed] [Google Scholar]