Abstract
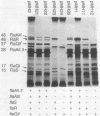
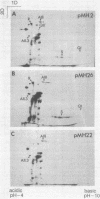
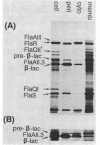
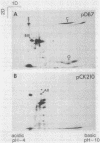
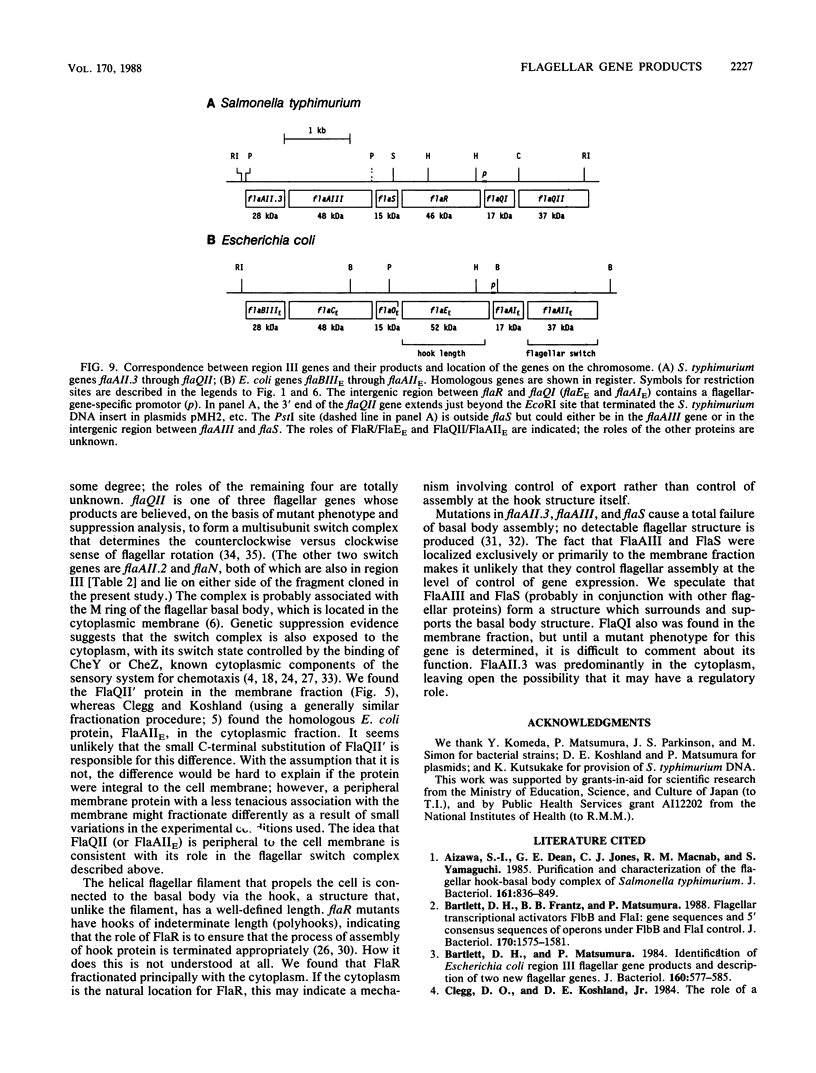
A portion of flagellar region III of the Salmonella typhimurium genome has been cloned and shown to contain six genes: flaAII.3, flaAIII, flaS, flaR, flaQI, and flaQII. Of these, all but flaQI were known to exist from mutant studies; the former flaQ has been renamed flaQII. The genes were shown by minicell analysis to encode proteins with apparent molecular masses of 28, 48, 15, 46, 17, and 37 kilodaltons, respectively. The presence of a flagellar-gene-specific promoter in the vicinity of flaQI was established by testing expression of the plasmid-encoded tetracycline resistance gene in artificial constructions. In minicell preparations, the flaAII.3 and flaR products were found principally in the cytoplasmic fraction; the rest were found principally in the membrane fraction. A comparison between the homologous genes of S. typhimurium and Escherichia coli confirmed that their genomic organizations were similar and that their products had similar molecular masses and isoelectric points.
Full text
PDF


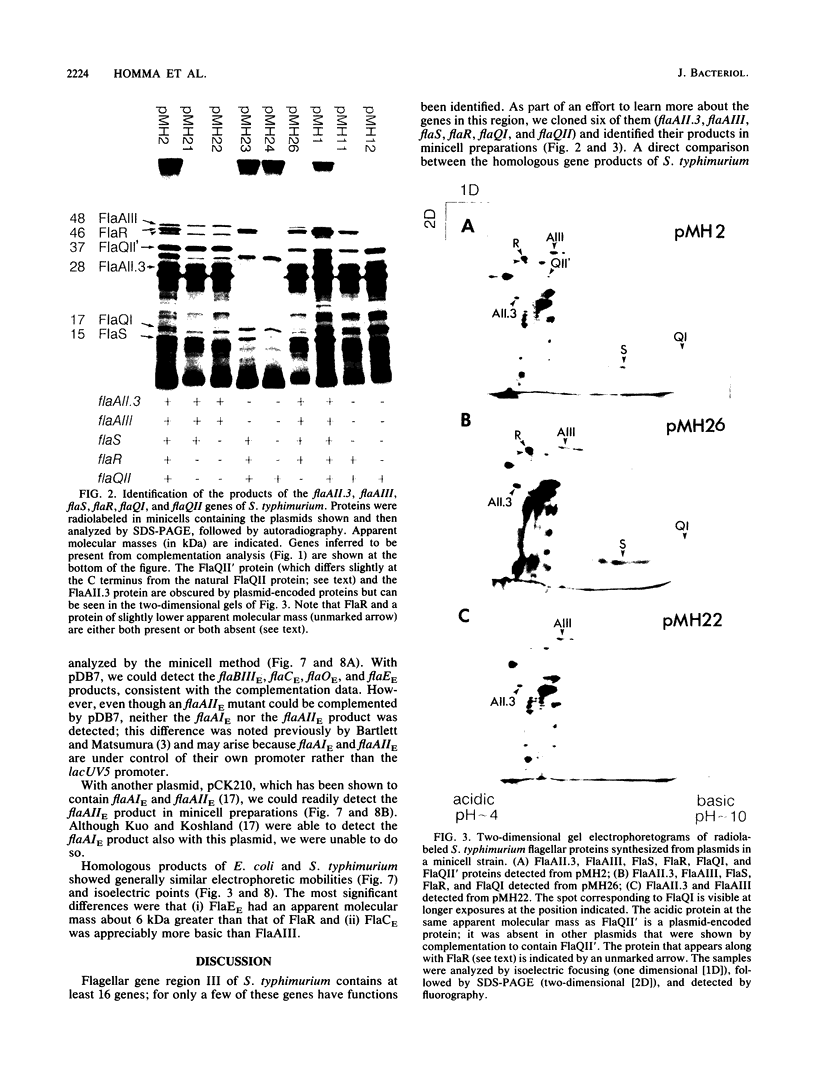
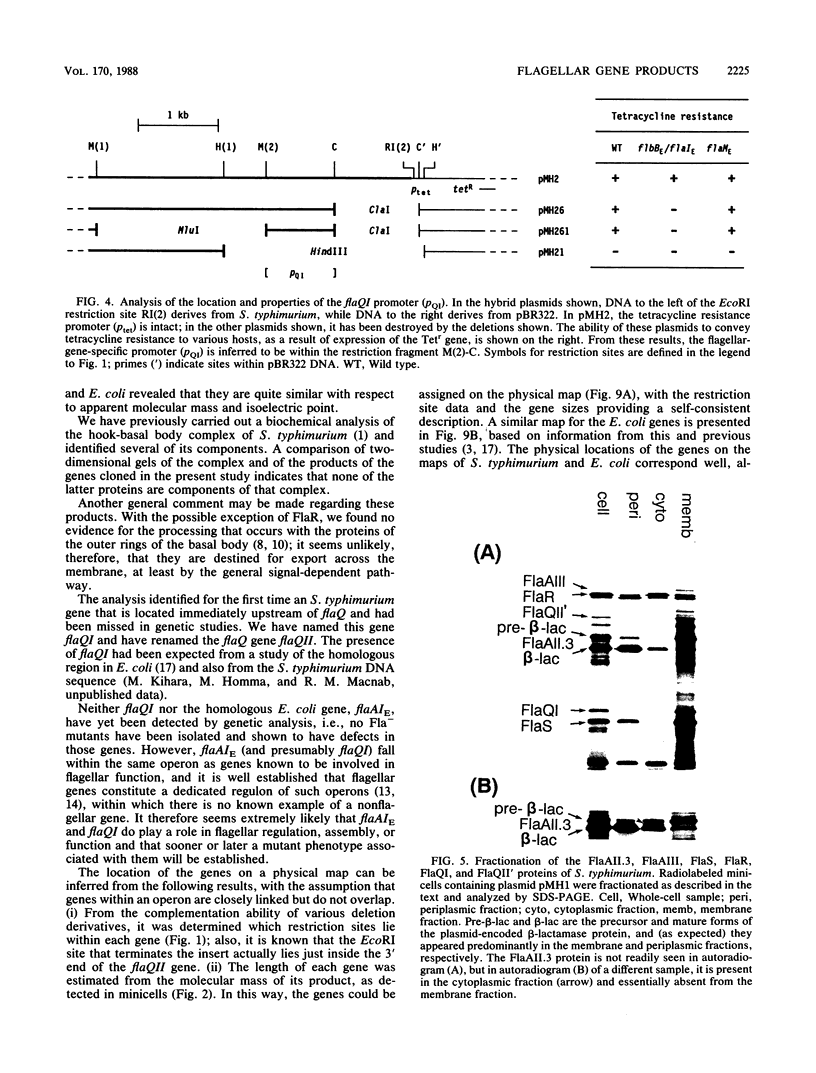
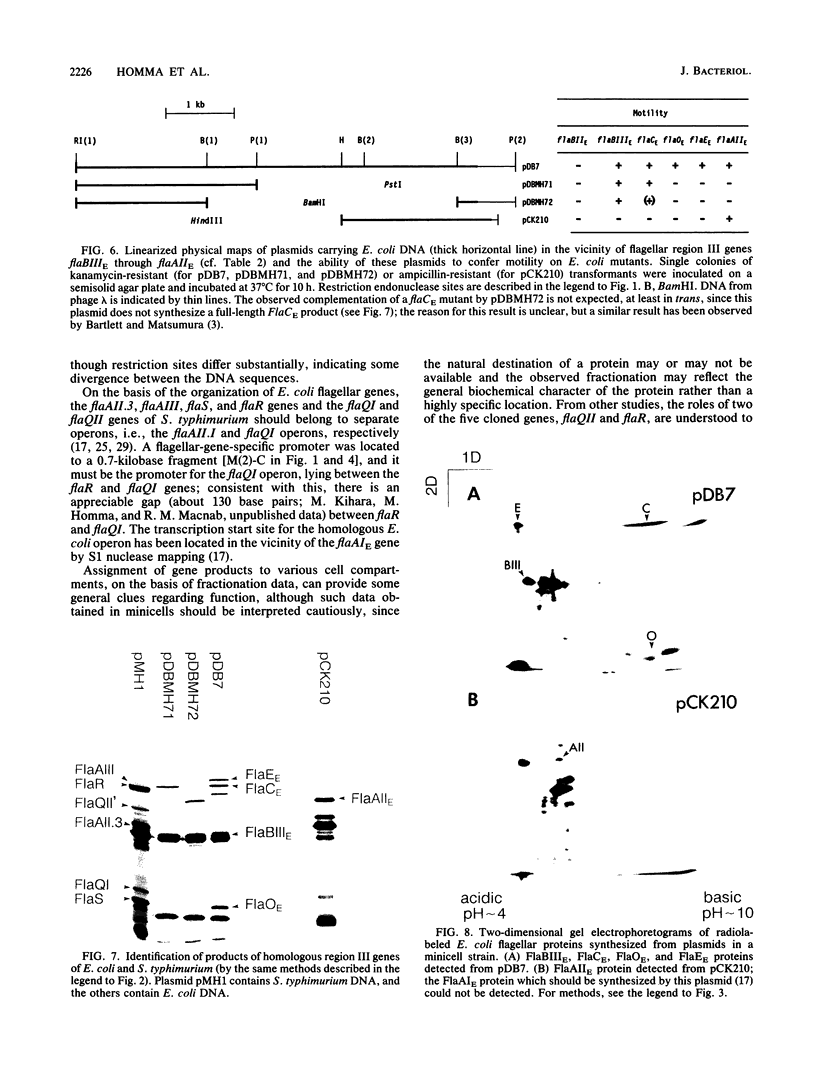

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aizawa S. I., Dean G. E., Jones C. J., Macnab R. M., Yamaguchi S. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J Bacteriol. 1985 Mar;161(3):836–849. doi: 10.1128/jb.161.3.836-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. H., Frantz B. B., Matsumura P. Flagellar transcriptional activators FlbB and FlaI: gene sequences and 5' consensus sequences of operons under FlbB and FlaI control. J Bacteriol. 1988 Apr;170(4):1575–1581. doi: 10.1128/jb.170.4.1575-1581.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett D. H., Matsumura P. Identification of Escherichia coli region III flagellar gene products and description of two new flagellar genes. J Bacteriol. 1984 Nov;160(2):577–585. doi: 10.1128/jb.160.2.577-585.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg D. O., Koshland D. E., Jr Identification of a bacterial sensing protein and effects of its elevated expression. J Bacteriol. 1985 Apr;162(1):398–405. doi: 10.1128/jb.162.1.398-405.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePamphilis M. L., Adler J. Attachment of flagellar basal bodies to the cell envelope: specific attachment to the outer, lipopolysaccharide membrane and the cyoplasmic membrane. J Bacteriol. 1971 Jan;105(1):396–407. doi: 10.1128/jb.105.1.396-407.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Aizawa S., Dean G. E., Macnab R. M. Identification of the M-ring protein of the flagellar motor of Salmonella typhimurium. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7483–7487. doi: 10.1073/pnas.84.21.7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Komeda Y., Iino T., Macnab R. M. The flaFIX gene product of Salmonella typhimurium is a flagellar basal body component with a signal peptide for export. J Bacteriol. 1987 Apr;169(4):1493–1498. doi: 10.1128/jb.169.4.1493-1498.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Kutsukake K., Iino T. Structural genes for flagellar hook-associated proteins in Salmonella typhimurium. J Bacteriol. 1985 Aug;163(2):464–471. doi: 10.1128/jb.163.2.464-471.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma M., Ohnishi K., Iino T., Macnab R. M. Identification of flagellar hook and basal body gene products (FlaFV, FlaFVI, FlaFVII and FlaFVIII) in Salmonella typhimurium. J Bacteriol. 1987 Aug;169(8):3617–3624. doi: 10.1128/jb.169.8.3617-3624.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iino T., Enomoto M. Genetical studies of non-flagellate mutants of Salmonella. J Gen Microbiol. 1966 Jun;43(3):315–327. doi: 10.1099/00221287-43-3-315. [DOI] [PubMed] [Google Scholar]
- Joys T. M. The covalent structure of the phase-1 flagellar filament protein of Salmonella typhimurium and its comparison with other flagellins. J Biol Chem. 1985 Dec 15;260(29):15758–15761. [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeda Y. Transcriptional control of flagellar genes in Escherichia coli K-12. J Bacteriol. 1986 Dec;168(3):1315–1318. doi: 10.1128/jb.168.3.1315-1318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H., Ozeki H. Deletion and amber mutants of fla loci in Escherichia coli K-12. Genetics. 1976 Nov;84(3):403–421. doi: 10.1093/genetics/84.3.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno R., Fujita H., Horiguchi T., Yamaguchi S. Precise position of the nml locus on the genetic map of Salmonella. J Gen Microbiol. 1976 Mar;93(1):182–184. doi: 10.1099/00221287-93-1-182. [DOI] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Roles of cheY and cheZ gene products in controlling flagellar rotation in bacterial chemotaxis of Escherichia coli. J Bacteriol. 1987 Mar;169(3):1307–1314. doi: 10.1128/jb.169.3.1307-1314.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo S. C., Koshland D. E., Jr Sequence of the flaA (cheC) locus of Escherichia coli and discovery of a new gene. J Bacteriol. 1986 Jun;166(3):1007–1012. doi: 10.1128/jb.166.3.1007-1012.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsukake K., Iino T., Komeda Y., Yamaguchi S. Functional homology of fla genes between Salmonella typhimurium and Escherichia coli. Mol Gen Genet. 1980 Apr;178(1):59–67. doi: 10.1007/BF00267213. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Nakao T., Iino T. A gene for DNA invertase and an invertible DNA in Escherichia coli K-12. Gene. 1985;34(2-3):343–350. doi: 10.1016/0378-1119(85)90143-x. [DOI] [PubMed] [Google Scholar]
- Matsumura P., Rydel J. J., Linzmeier R., Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984 Oct;160(1):36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Parker S. R., Talbert P. B., Houts S. E. Interactions between chemotaxis genes and flagellar genes in Escherichia coli. J Bacteriol. 1983 Jul;155(1):265–274. doi: 10.1128/jb.155.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson-Delafield J., Martinez R. J., Stocker B. A., Yamaguchi S. A new fla gene in Salmonella typhimurium--flaR--and its mutant phenotype-superhooks. Arch Mikrobiol. 1973 Mar 26;90(2):107–120. doi: 10.1007/BF00414513. [DOI] [PubMed] [Google Scholar]
- Ravid S., Matsumura P., Eisenbach M. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7157–7161. doi: 10.1073/pnas.83.19.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M. R., Simon M. I. Flagellar assembly mutants in Escherichia coli. J Bacteriol. 1972 Nov;112(2):986–993. doi: 10.1128/jb.112.2.986-993.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M., Simon M. Positioning flagellar genes in Escherichia coli by deletion analysis. J Bacteriol. 1974 Jan;117(1):73–79. doi: 10.1128/jb.117.1.73-79.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Iino T., Horiguchi T., Yamaguchi S. Incomplete flagellar structures in nonflagellate mutants of Salmonella typhimurium. J Bacteriol. 1978 Feb;133(2):904–915. doi: 10.1128/jb.133.2.904-915.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Iino T. Role of the flaR gene in flagellar hook formation in Salmonella spp. J Bacteriol. 1981 Dec;148(3):973–979. doi: 10.1128/jb.148.3.973-979.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Komeda Y. Incomplete flagellar structures in Escherichia coli mutants. J Bacteriol. 1981 Feb;145(2):1036–1041. doi: 10.1128/jb.145.2.1036-1041.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Conley M. P., Kramer T. J., Berg H. C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987 May;169(5):1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Aizawa S., Kihara M., Isomura M., Jones C. J., Macnab R. M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986 Dec;168(3):1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S., Fujita H., Ishihara A., Aizawa S., Macnab R. M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986 Apr;166(1):187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]