Abstract
We tested the ability of recombinant hMutSα (hMSH2/hMSH6) and hMutSβ (hMSH2/hMSH3) heterodimers to complement the mismatch repair defect of HEC59, a human cancer cell line whose extracts lack all three MutS homologues. Although repair of both base/base mispairs and insertion–deletion loops was restored by hMutSα, only the latter substrates were addressed in extracts supplemented with hMutSβ. hMutSα was also able to complement a defect in the repair of base/base mispairs in CHO R and HL60R cell extracts. In these cells, methotrexate-induced amplification of the dihydrofolate reductase (DHFR) locus, which also contains the MSH3 gene, led to an overexpression of MSH3 and thus to a dramatic change in the relative levels of MutSα and MutSβ. As a rule, MSH2 is primarily complexed with MSH6. MutSα is thus relatively abundant in mammalian cell extracts, whereas MutSβ levels are generally low. In contrast, in cells that overexpress MSH3, the available MSH2 protein is sequestered predominantly into MutSβ. This leads to degradation of the partnerless MSH6 and depletion of MutSα. CHO R and HL60R cells therefore lack correction of base/base mispairs, whereas loop repair is maintained by MutSβ. Consequently, frameshift mutations in CHO R are rare, whereas transitions and transversions are acquired at a rate two orders of magnitude above background. Our data thus support and extend the findings of Drummond et al. [Drummond, J. T., Genschel, J., Wolf, E. & Modrich, P. (1997) Proc. Natl. Acad. Sci. USA 94, 10144–10149] and demonstrate that mismatch repair deficiency can arise not only through mutation or transcriptional silencing of a mismatch repair gene, but also as a result of imbalance in the relative amounts of the MSH3 and MSH6 proteins.
In eukaryotes, mismatch recognition is mediated predominantly by MutSα, a heterodimer of MSH2 and MSH6, two homologues of the bacterial mismatch-binding protein MutS (1–5). Surprisingly, although these polypeptides function in the repair of mismatches and small insertion/deletion loops (IDLs) as a complex, the phenotype of cells mutated in the two respective genes is different (2, 6). Thus, although human cells mutated in hMSH2 are deficient in the correction of both base/base mispairs and IDLs, and consequently display instability of mono- and dinucleotide microsatellite repeats (7, 8), cells lacking hMSH6 [also known as p160 (2) or GTBP (1)] were shown to be deficient principally in the correction of base/base mismatches (2) and to display an instability of microsatellite motifs consisting of mononucleotide runs only (5). Moreover, extracts of the hMSH6-deficient cells retained a residual capacity for IDL repair (2), and thus it was postulated that, in these cells, hMSH2 must be mediating loop recognition either alone, or together with another MutS homolog (9). This prediction was soon confirmed by genetic and biochemical experiments carried out in Saccharomyces cerevisiae, which implicated MSH3 as the alternative partner of MSH2 (5). Subsequent studies from our laboratory showed that hMSH3 and hMSH2 formed a heterodimer, hMutSβ, which could bind IDLs but not base/base mismatches in an in vitro assay (10). Support for the involvement of hMSH3 in IDL repair also came from experiments that demonstrated that transfer of chromosome 5, which contains the hMSH3 gene, into an endometrial cancer line HHUA (mutated in both hMSH6 and hMSH3) resulted in the expression of hMSH3 and in the correction of at least a subset of IDL-containing substrates (11). With the above evidence in mind, we set out to obtain direct biochemical proof of the involvement of MutSβ in postreplicative mismatch correction.
The functional redundancy of MutSα and MutSβ in IDL repair could have several consequences. First, the loss of MSH6 should lead to a mutator phenotype through the lack of correction of base/base mispairs, whereas the repair of IDLs, mediated by MutSβ, ought to maintain microsatellite instability at low levels. As noted above, this was found to be the case in cell lines mutated in both alleles of the hMSH6 gene (6), as well as in mouse MSH6−/− lines (12). Second, the loss of MSH3 would be expected to have only a small effect on global mismatch repair. Indeed, this appears to be the case both in yeast, where msh3 mutants displayed only a limited dinucleotide repeat instability (13), and in humans, where bone marrow cells from several patients with hematological malignancies could be shown to express extremely low levels of hMSH3 mRNA (14), yet failed to display any phenotype that could be associated with the lack of mismatch repair (T. Shimada and M. Ikejima, personal communication). Third, that MSH6 and MSH3 compete for MSH2 predicts that elevated expression of either protein would result in the “squelching” of MSH2 in favor of one or the other heterodimer. The consequences of disregulation of MSH6 or MSH3 would, however, be dramatically different. Thus, whereas elevated expression of MSH6 would be expected to have only a small effect on global mismatch repair (similar to the loss of MSH3), overexpression of MSH3 should result in the reduction in the levels of MutSα and therefore in the reduction of the efficiency of repair of base/base mismatches.
In an attempt to test this last hypothesis, we set out to study the mismatch repair capacity of cells overexpressing MSH3. Because the MSH3 gene was shown to be divergently transcribed from the dihydrofolate reductase (DHFR) promoter in both rodents (15, 16) and man (17), we decided to examine cell lines with an amplified DHFR locus. One candidate line was generated by Drobetsky and Meuth (18), who treated Chinese hamster ovary (CHO) cells with increasing amounts of the cytotoxic drugs 5-fluoro-2′-deoxyuridine (FdU) and methotrexate (MTX), in the hope of isolating cells with a mutator phenotype and thus of identifying novel genes responsible for the maintenance of genomic stability. The rationale of these experiments lay in the expectation that mutators might adapt more rapidly to the selective pressure of their environment. After both treatments, several FdU- and MTX-resistant clones were indeed identified. Although it became apparent that their resistance to the drug was achieved primarily through elevated expression of the target enzymes, thymidylate synthase and DHFR, respectively, the MTX-resistant clones also displayed elevated mutation rates to ouabain, 6-thioguanine, and emetine resistance. In a follow-up study (19), the MtxR cells (referred to as CHO R in the present study) were shown to mutate to 6-thioguanine resistance at a rate of 1.7 × 10−6, which was 100-fold higher than in the wild-type cells. Sequence analysis of the mutated hprt gene revealed that although the spectrum of mutations in wild-type cells consisted predominantly of transversions and frameshifts, the mutations found in the CHO R clone were exclusively transitions and transversions. Although the reason underlying this phenotype was unclear at that time, the authors suggested that either replication fidelity was directly affected in this clone, or that error correction mechanisms, such as proofreading or mismatch correction, have become less efficient. As the DHFR locus was shown to be amplified more than 500-fold in the CHO R line (19), we predicted, based on our current knowledge, that this amplification resulted also in an overexpression of MSH3, which led, in turn, to an elevation of MutSβ levels at the expense of MutSα. The mutator phenotype of the CHO R cells was thus predicted to be a result of a deficiency in the repair of base/base mismatches. To provide supporting evidence for this hypothesis, we studied the substrate specificity of mismatch repair in extracts of the CHO R cells. We also included in this investigation the human leukemia cells HL60R, in which the DHFR locus was shown to be amplified 200-fold (17), and which we anticipated to have a phenotype similar to that of the CHO R cells. We show that amplification of the DHFR locus in the two cell lines has indeed resulted in an overexpression of the MSH3 gene and that this severely affected the ratio of MutSα to MutSβ. As a result, these cells are deficient in the repair of base/base mismatches and have a strong mutator phenotype. Our data support the findings of a similar, independent study by Drummond et al. (20) and extend them also to rodent cells.
MATERIALS AND METHODS
Expression and Purification of hMutSα and hMutSβ.
The baculovirus vectors carrying cDNA inserts encoding the hMSH6, hMSH2, and hMSH3 proteins (10) were used to infect cultures of Sf9 cells (GIBCO). Although single infections with the hMSH6 and hMSH3 vectors failed to yield reasonable amounts of the respective recombinant proteins, coinfection with hMSH2 and hMSH6 or with hMSH2 and hMSH3 viral vectors resulted, respectively, in the expression of hMutSα and hMutSβ in high yields. The procedure for protein recovery and purification was described previously (10).
Cell Lines and Extract Preparation.
The HEC59 line was a kind gift of Thomas Kunkel (National Institute on Environmental Health Sciences, Research Triangle Park, NC). These cells were grown in DMEM/F12 medium, 1:1, supplemented with 20% fetal bovine serum (GIBCO). The CHO and CHO R (MtxR) lines were kindly provided by Mark Meuth (University of Utah). They were grown in α-MEM, supplemented with 10% fetal bovine serum (GIBCO). The HL60 and HL60R lines were a kind gift of Takashi Shimada (Nippon Medical School, Tokyo). They were grown in RPMI 1640 medium supplemented with 20% fetal bovine serum (GIBCO). HeLa cells were grown in DMEM, supplemented with 10% fetal bovine serum (GIBCO). The culture media of the methotrexate-resistant lines were supplemented with 1 μM MTX. The cytoplasmic extracts were prepared as described (21), without modification. The typical protein concentrations were around 10 μg/μl.
Antibodies.
The anti-hMSH3 polyclonal antibodies were a kind gift of Takashi Shimada. The rabbit anti-hMSH2 polyclonal serum was as described previously (1). The mouse anti-hMSH6 mAbs 66H6 and 2D4, both IgGγ1, were raised against the full-length recombinant hMSH6 protein. Female balb/c mice were immunized with four intraperitoneal and one intravenal injection. The spleen cells were then fused with the myeloma line P3x63 Ag8.653. Hybrids were selected with ELISA and Western blots. They were then cloned by limiting dilution, and the individual clones were screened again as above. The selected mAb-secreting lines were adapted to grow in roller bottles at low percentage (1%) of fetal calf serum. The antibodies were purified from the culture medium by Gamma-plus Protein G Sepharose (Pierce).
Tryptic Digests of hMSH2 and hMSH6.
The purified human hMSH2/hMSH6 heterodimer (500 ng) was subjected to a partial proteolysis with 0, 5, 10, 50, 100, or 600 ng of trypsin as described previously (22). The fragments were separated on a 7.5% denaturing polyacrylamide gel and electrotransferred onto nitrocellulose membranes. Western blotting was carried out as described below, using rabbit polyclonal anti-hMSH2 and anti-hMSH6 antisera diluted 1:1,000 (1).
Western Blotting and Bandshift Experiments.
The Western blotting procedure employed in this study was described previously (1), using the rabbit polyclonal anti-hMSH2 and the mouse monoclonal anti-hMSH6 antibody 2D4. Bandshift assays were carried out as described (10), using either a 34-mer probe G/T, or a 34/36-mer carrying a loop of two extrahelical nucleotides (heteroduplex E in ref. 10). In the supershift experiments, 1 μg of the monoclonal hMSH6 antibody 66H6 was added to the reaction mixtures at the beginning of the incubation period.
Mismatch Repair Assays.
The efficiency of cytoplasmic extracts in repairing DNA mismatches or loops was tested as described previously (21, 23). M13 mp2 heteroduplexes containing either a G/G mispair or a single loop of two extrahelical nucleotides were incubated with the cell extracts as described (21, 23). The DNA was then purified, electroporated into a mutS strain of Escherichia coli, and plated along with the α-complementation strain CSH50, isopropyl β-d-thiogalactoside, and 5-bromo-4-chloro-3-indolyl β-d-galactoside. If no repair occurred, a high percentage of mixed plaques, containing both blue and colorless progeny, was observed. Reduction in the percentage of mixed plaques and a concomitant increase in single-color plaques were indicative of repair. In the complementation studies, the assays were carried out as described above, except that the extracts (50 μg) were supplemented with purified recombinant hMutSα or hMutSβ (0.1 μg each). Repair efficiency (%) = 100 × [1 − (% mixed plaques in extract-treated sample)/(% mixed plaques in extract-untreated sample)].
RESULTS
Recombinant hMutSα and hMutSβ Complement Mismatch Repair Defects of HEC59 Extracts.
Although hMutSα purified from human (HeLa) cells was shown to restore repair of base/base mismatches and IDLs in extracts of cells lacking hMSH2 or hMSH6 (2), no direct biochemical proof of the involvement of hMutSβ in IDL repair was available at the start of this study. Therefore we expressed both these heterodimeric factors in the baculovirus system (10) and tested their ability to complement extracts of mismatch repair-deficient cells. We chose the human endometrial cancer line HEC59 for these control experiments, because it has mutations in both alleles of hMSH2 (24) and therefore lacks both mismatch- and IDL-binding activity (Fig. 1 Center). Moreover, the absence of hMSH2 in this line resulted in a destabilization of hMSH6 (1) (see also Fig. 4), and hMSH3 was undetectable by Western blot under standard conditions (Fig. 2). HEC59 extracts thus effectively lack all three MutS homologs and are therefore ideally suited for complementation studies.
Figure 1.
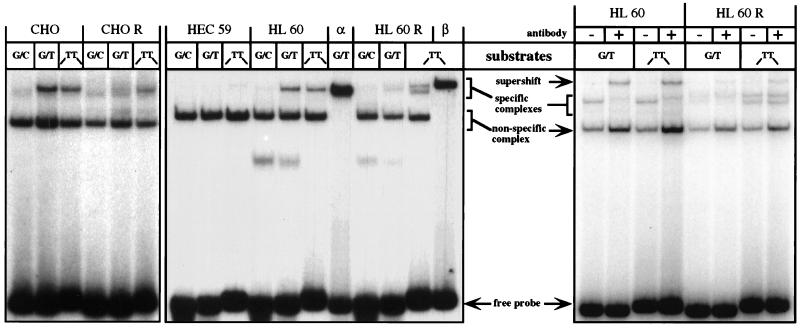
Mismatch- and IDL-binding activities in extracts of CHO, CHO R, HEC59, HL60, and HL60R cells. The extracts were incubated with radioactively labeled oligonucleotide duplexes (see Materials and Methods) either perfectly complementary (G/C) or containing a single mispair (G/T) or an IDL containing two extrahelical thymines (-TT-) (10). (Left and Center) The bandshift experiments were carried out with cytoplasmic extracts of CHO, CHO R, HEC59, HL60, and HL60R cells. The electrophoretic mobility of the protein/DNA complexes was compared with those formed upon incubation of the oligonucleotide substrates with the purified recombinant hMutSα (lane α) or hMutSβ (lane β). Note that, unlike the purified recombinant hMSH3/hMSH2 heterodimer, the hMutSβ present in human cell extracts gave rise to two distinct protein/DNA complexes (lane HL60R/TT). Neither of these complexes comigrates with hMutSα-bound substrates (e.g., lane HL60R/G/T) (see also text). (Right) Supershift experiments using the anti-hMSH6 mAb 66H6. Addition of the mAb to the binding mixtures containing extracts of HL60 or HL60R cells retarded the mobility of the oligonucleotide probes bound by hMutSα. The figure is an autoradiograph of a nondenaturing 6% polyacrylamide gel run in TAE buffer.
Figure 4.
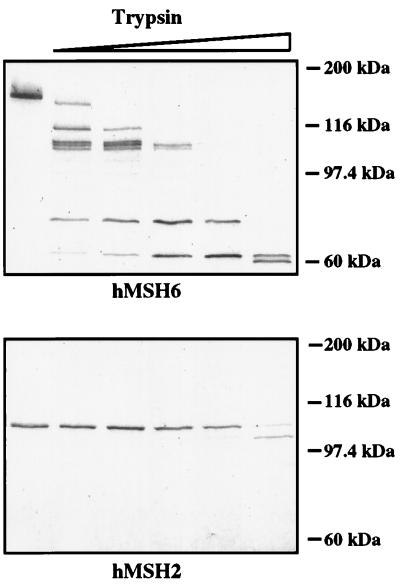
Partial proteolysis of hMSH2 and hMSH6 within the context of hMutSα. Purified recombinant hMutSα was incubated (from left to right) with 0, 5, 10, 50, 100, and 600 ng trypsin as described in Materials and Methods. The products were separated on SDS/PAGE, transferred onto nitrocellulose membranes, and visualized with polyclonal anti-hMSH2 (Lower) and anti-hMSH6 (Upper) antisera.
Figure 2.
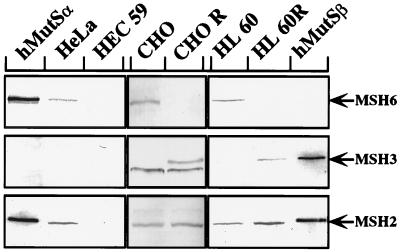
Immunoblot of MSH2, MSH3, and MSH6 polypeptides present in extracts of HeLa, CHO, CHO R, HEC59, HL60, and HL60R cell lines. The band migrating just below MSH3 in the hamster cell extracts is caused by a protein cross-reacting with the polyclonal anti-hMSH3 antiserum. The figure is a Western blot of cytoplasmic extracts stained with rabbit polyclonal antisera raised against the three proteins. The extracts were prepared as described (21), and 20 μg per lane were loaded on a 7.5% denaturing SDS-polyacrylamide gel. The proteins were transferred onto nitrocellulose membranes, which were then stained with the indicated antisera. hMutSα and hMutSβ, purified recombinant heterodimers included for reference.
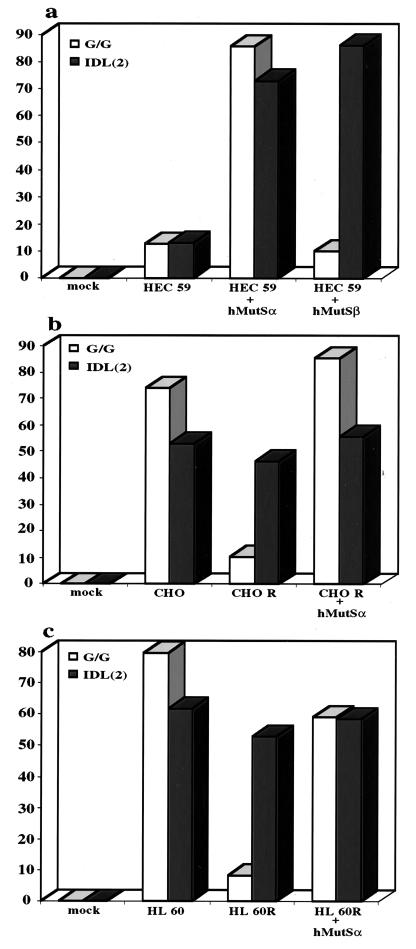
Using an in vitro mismatch repair assay (23), we could show that HEC59 extracts were deficient in the repair of a G/G mismatch and an IDL containing two extrahelical nucleotides (Fig. 3a). Addition of recombinant hMutSα to HEC59 extracts could restore the repair of both these substrates, whereas only the IDL substrate was corrected upon the addition of hMutSβ (Fig. 3a). These results confirm the prediction (5, 10, 11) that hMutSβ does not play a major role in the correction of base/base mismatches, at least as measured by our in vitro assay, and imply that the residual two-nucleotide-loop repair observed in extracts of hMSH6-deficient DLD1 (HCT15) cells (2) could have been mediated by this factor. However, available experimental evidence suggests that hMutSβ takes part in the repair of only a subset of IDLs (11), and it is to be expected that, under normal circumstances, the repair of base/base mispairs and small IDLs in eukaryotic cells would be mediated predominantly by MutSα. This prediction is based on the finding that whereas MutSα is abundant in all mismatch repair-proficient cells examined to date (Fig. 2 and our unpublished data; see also refs. 2 and 20), the expression of MSH3 is generally extremely low (Fig. 2).
Figure 3.
Efficiency of mismatch correction in extracts of HEC59, CHO, CHO R, HL60, and HL60R cell lines. The extracts were supplemented with purified recombinant hMutSα or hMutSβ as indicated below the columns (see also Materials and Methods). (a) Repair efficiency of the G/G mispair and of the two nucleotide IDL in HEC59 extracts supplemented or not with hMutSα or hMutSβ. (b) As in a, except that CHO and CHO R extracts were used. (c) As in b, except that HL60 and HL60R extracts were used.
Extracts of MTX-Resistant Cells Are Depleted of MutSα, but Contain Abundant Levels of IDL-Binding Activity.
In bandshift experiments, which were carried out by using extracts of CHO and HL60 cells, an activity binding both the G/T mismatch and the IDL is clearly detectable (Fig. 1) and can be seen to comigrate with the specific band produced by purified recombinant hMutSα. In contrast, the CHO R and HL60R extracts were almost depleted of the G/T-binding activity. Instead, they contained a significant amount of IDL-specific complex, which was distinct from hMutSα. In HL60R extracts, two IDL-specific bands could be seen: one comigrating with the recombinant hMutSβ complex, the other of somewhat higher mobility. We believe that both these species represent the hMSH2/hMSH3 heterodimer, the observed differences being most likely caused by alternative, posttranslational modification of hMSH3. In contrast to the human cells, the complex of the IDL probe with MutSβ present in the extracts of the hamster CHO R cells appeared as a single band (Fig. 1 Left).
Additional evidence concerning the identity of the mismatch-binding species was obtained in a series of supershift experiments carried out with the monoclonal anti-hMSH6 antibody 66H6, which recognizes the native protein (see Materials and Methods). As shown in Fig. 1 (Right), when this antibody was added to binding mixtures containing extracts of HL60 cells, the mismatch-specific bands were supershifted. This indicates that the protein factor bound to the G/T mispair and to the two nucleotide IDL contained hMSH6. In contrast, extracts of the HL60R line contained only a small amount of hMutSα, which migrated in the bandshift experiment between the two complexes because of hMutSβ (Fig. 1 Center). Whereas this band was duly shifted to lower mobility after addition of the 66H6 antibody (lane G/T +), the predominant IDL-binding activity in these extracts (lane TT +) was not affected. Furthermore, because the intensity of both bands was decreased by the addition of polyclonal hMSH3 antisera (data not shown), we presume that this loop-binding factor is hMutSβ.
The MSH3 Gene Coamplifies with the DHFR Locus in MTX-Resistant Cells.
In extracts of mismatch repair-proficient cells, exemplified here by the hamster CHO line and by the human leukemia line HL60, MSH2 and MSH6 are present in approximately equal amounts (Fig. 2). As already discussed, methotrexate treatment of the CHO and HL60 cell cultures led to a several hundredfold amplification of the DHFR locus. As mentioned above, the MSH3 gene is divergently transcribed from the same promoter/enhancer both in the human (17) and in the rodent (16, 25) genomes, and the amplification of the DHFR locus resulted in an overexpression of MSH3, which became easily detectable in the CHO R and HL60R extracts by Western blotting (Fig. 2). Northern blot analysis revealed that although the amounts of hMSH6 mRNA were comparable in CHO and CHO R cells, the quantity of hMSH3 mRNA increased from barely detectable levels in CHO cells to an amount similar to that of hMSH6 mRNA in the CHO R line (data not shown). This evidence thus not only confirms that the expression of the MSH3 gene is greatly increased in the latter cells, it demonstrates that the MSH6 gene is not shut off in the methotrexate-resistant cells.
MSH3 Overexpression Results in the Loss of MSH6 and MutSα.
As shown in Fig. 2, the CHO R and HL60R extracts appeared to be devoid of MSH6. We noted that this polypeptide was also barely detectable in extracts of LoVo (1) and HEC59 cells (Fig. 2), which lack hMSH2, and we suspected that the reason underlying the degradation of hMSH6 in these lines is that this protein is susceptible to proteolysis in the absence of its partner, hMSH2. Our present data suggest that a similar situation also exists in CHO R and HL60R extracts, albeit with an important difference: namely, that MSH6 is without a partner in these extracts not because of MSH2 having been mutated or transcriptionally silenced, but rather because of its having been sequestered into MutSβ. This hypothesis is rather difficult to substantiate experimentally not only in vivo, but also in vitro, because the MSH6 protein is, unlike MSH2, difficult to express in large amounts. However, when we examined the sensitivity of the hMutSα heterodimer to trypsin, hMSH6 could be shown to be substantially more prone to degradation by this enzyme than its cognate partner (Fig. 4). When this evidence is considered in light of the fact that hMSH6 mRNA levels were similar in both CHO and CHO R cells (see preceding section) and that hMSH6 alone could not be overexpressed from a baculovirus vector (10), the hypothesis that this protein is prone to proteolytic degradation seems plausible.
Depletion of MutSα Results in the Loss of Correction of Base/Base Mispairs.
We decided to test whether the reduction of G/T mismatch-binding activity was accompanied also by mismatch repair deficiency. Extracts of the two matched pairs of cell lines, CHO and CHO R, as well as HL60 and HL60R were therefore tested for their ability to correct a heteroduplex containing either a base/base mismatch or an IDL of two extrahelical nucleotides. As shown in Fig. 3 b and c, the CHO and HL60 extracts were proficient in the repair of both these substrates, whereas in CHO R and HL60R extracts only the IDL was corrected. The addition of purified recombinant hMutSα to the CHO R (Fig. 3b) and HL60R (Fig. 3c) extracts resulted in restoration of base/base mismatch repair, which confirmed that the defect in these cells rests in the lack of a functional MSH2/MSH6 heterodimer. Purified hMSH6 alone failed to complement the mismatch repair defect in the HL60R extracts, as it appears to be unable to displace hMSH3 from a preformed complex with hMSH2 (data not shown). An interesting observation concerns the fact that the hMSH2/hMSH6 heterodimer also complemented extracts of the CHO R hamster cells (Fig. 3b). The ability of human mismatch repair proteins to interact with the other members of the rodent mismatch repair machinery indicates a high degree of conservation and is reminiscent of nucleotide excision repair genes, many of which were cloned thanks to their ability to cross-complement defects of UV-sensitive CHO lines (26).
DISCUSSION
Redundancy of Function of MutSα and MutSβ.
The finding that HCT15 (DLD1) and MT1 cells, which have mutations in both alleles of the hMSH6 gene, are deficient in the repair of base/base mismatches, while retaining some ability to correct IDLs (2), led to the prediction that hMSH2 must have a partner other than hMSH6, together with which it can function in loop recognition. Genetic experiments carried out with yeast (5, 13) and human (11) cells mutated in the MSH3, MSH6, or both identified this gene as MSH3 and implied that MSH3 and MSH6 both can play a part in IDL repair. In support of these predictions we were able to demonstrate (10) that recombinant hMutSα and hMutSβ both were able to bind to oligonucleotide substrates containing extrahelical nucleotides. Our present results show that the two heterodimers are also capable of initiating the strand-directed IDL correction process in an in vitro assay (Fig. 3). However, Western blot experiments indicate that extracts of mammalian cells in culture generally contain only very low amounts of MSH3, despite the fact that the MSH3 gene is expressed and its mRNA can be shown to be present in a quantity similar to that of the DHFR message (16, 27), which is divergently transcribed from the same promoter (16, 17, 25); clearly, this evidence would argue against a major role of MutSβ in loop repair in normal-growing cells. It was therefore puzzling to discover that in HCT15 (DLD1) cells, IDL repair could be detected in an in vitro repair assay (2), despite the complete lack of hMutSα and the low levels of hMutSβ. Our present results help to explain these apparently discordant findings. Under normal growth conditions, all three MutS homologues are expressed, but MSH2 forms a heterodimer preferentially with MSH6, because both of these proteins are expressed very efficiently. Any MSH3 that may be translated either fails to find available free MSH2, or it becomes degraded in the absence of its cognate partner. In the case where MSH3 is overexpressed, such as in the lines described above, the situation is reversed: MSH2 complexes predominantly with MSH3, and MSH6 is degraded (Fig. 4; see also ref. 10). A similar situation would arise when MSH6 is absent, either because of a mutation in the MSH6 gene or possibly because of transcriptional silencing. In this case, MSH2 would recruit all available MSH3 into MutSβ, with the result that the genomes of these cells, despite being subjected to a considerable mutagenic load, would at least stay free of polymerase slippage errors in repeated sequence motifs and, thus, of frameshift mutations.
We have at this time no direct evidence in support of the hypothesis that the partnerless MSH3 or MSH6 are proteolytically degraded. However, we did show previously that expression of the two human proteins in the baculovirus system in the absence of hMSH2 yielded either very low yields of product or material that was largely insoluble (10); coinfection with viruses expressing hMSH2 and one of its partners resulted in the expression of large amounts of the soluble heterodimers. Moreover, purified hMSH6 in the context of the heterodimer is substantially more prone to degradation with trypsin than hMSH2 (Fig. 4), implying that its structure is more accessible to proteolytic enzymes.
We cannot at present exclude the possibility that the expression of the MSH3 gene is subject to complex regulation and that, rather than playing merely a backup role, MSH3 becomes highly expressed in certain tissues or cell types, where it fulfills a specific role that has so far escaped our attention. However, we consider this possibility unlikely, because the sequences known to control the expression of both the MSH3 and the DHFR genes appear to fall into the category of typical housekeeping gene promoters (16, 27).
Relevance of the Above Findings to Human Cancer.
Relapse after cancer chemotherapy is mostly a result of outgrowth of drug-resistant tumor cells. Drug resistance can take many forms, the most common of which involves the overexpression of the P-glycoprotein (p170) encoded by the MDR1 (multiple drug-resistance) gene, the mutation of the gene encoding the target protein such that the mutant form no longer recognizes the drug, or the amplification of the genomic locus encoding the target protein (see ref. 28 for review). MTX is highly effective in the treatment of childhood acute lymphocytic leukemia, but it is also used routinely in the clinic to treat other tumors, such as osteosarcoma and breast cancer. It is also used in the treatment of some autoimmune diseases such as refractory rheumatoid arthritis, psoriasis, and for the prevention of graft-versus-host disease in transplant patients (see ref. 29 for review). Resistance to MTX caused by amplification of the DHFR gene is frequently encountered in the clinic (30), and it is possible that even a small increase in hMSH3 levels could significantly alter the relative intracellular concentrations of hMutSα and hMutSβ and thus lower mismatch repair efficiency. Importantly, because amplification of the DHFR gene under MTX-selective pressure was reported to be exacerbated by induction of expression of the c-myc oncogene (31), which is overexpressed frequently in tumors, it is conceivable that the amplification process might be facilitated in cancer. This hypothesis is supported by numerous reports showing that increased expression (whether through genomic amplification or transcriptional activation) of genes involved in drug resistance (including DHFR) appears to be common in tumors lacking the product of the Rb gene, overproducing cyclin D1, or with altered p53 (see ref. 30 for review). Because p53 mutations are found in around 50% of all cancers (32), it is possible that MTX therapy might select for clones with diminished mismatch repair efficiency whose mutator phenotype would accelerate the acquisition of mutations in other tumor suppressor genes and oncogenes that are necessary for the progression of malignant disease. Moreover, such cancers would become rapidly refractory to chemotherapy, as witnessed by the increased tolerance of mismatch repair-deficient cells to alkylating agents (33), as well as to other DNA-modifying drugs such as cis-platin (34). Anecdotally, alkylating agents are often used together with MTX in chemotherapy cocktails, and it is possible that the two classes of drugs might act synergistically to select for mismatch repair-deficient and, therefore, drug-resistant clones.
While this work was in progress, Drummond et al. (20) presented data that showed that hMutSβ purified from HL60R cells could restore IDL repair to extracts of mismatch repair-deficient LoVo cells. Moreover, they demonstrated that HL60R cells have a mutator phenotype comparable to that of the CHO R line generated by Drobetsky and Meuth (18). These results, together with the data presented above, provide new evidence that mismatch repair deficiency can arise not only as the result of mutations or because of transcriptional down-regulation of mismatch repair genes (see ref. 35 for review), but that imbalance in expression of the individual components of the mismatch repair machinery also can result in a severe malfunction of the repair process.
Acknowledgments
We express our gratitude to Christine Hemmerle for expert technical assistance, to Drs. Takashi Shimada and Myioko Ikejima (Nippon Medical School, Tokyo) for the gift of the hMSH3-expressing baculovirus vector, the anti-hMSH3 polyclonal antiserum, and the HL60 and HL60R lines, and to Dr. Mark Meuth for the gift of the CHO and CHO R lines. We are also grateful to Tom Kunkel (National Institute on Environmental Health Sciences) for the gift of the HEC59 line and of the mutant and wild-type M13 mp2 phage, and to Primo Schär for discussions and for critical reading of the manuscript. Generous financial support of the Swiss National Research Foundation to J.J. is gratefully acknowledged.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: IDL, insertion/deletion loop; CHO, Chinese hamster ovary; MTX, methotrexate; DHFR, dihydrofolate reductase.
References
- 1.Palombo F, Gallinari P, Iaccarino I, Lettieri T, Hughes M, D’Arrigo A, Truong O, Hsuan J J, Jiricny J. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 2.Drummond J T, Li G M, Longley M J, Modrich P. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 3.Iaccarino I, Palombo F, Drummond J, Totty N F, Hsuan J J, Modrich P, Jiricny J. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 4.Acharya S, Wilson T, Gradia S, Kane M F, Guerrette S, Marsischky G T, Fishel R. Proc Natl Acad Sci USA. 1996;93:13629–13634. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 6.Papadopoulos N, Nicolaides N C, Liu B, Parsons R, Lengauer C, Palombo F, D’Arrigo A, Markowitz S, Willson J K, Kinzler K W, et al. Science. 1995;268:1915–1917. doi: 10.1126/science.7604266. [DOI] [PubMed] [Google Scholar]
- 7.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom Lahti M, et al. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 8.Fishel R, Lescoe M K, Rao M R, Copeland N G, Jenkins N A, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 9.Karran P. Science. 1995;268:1857–1858. doi: 10.1126/science.7604258. [DOI] [PubMed] [Google Scholar]
- 10.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 11.Risinger J I, Umar A, Boyd J, Berchuck A, Kunkel T A, Barrett J C. Nat Genet. 1996;14:102–105. doi: 10.1038/ng0996-102. [DOI] [PubMed] [Google Scholar]
- 12.de Wind N, Dekker M, Berns A, Radman M, te Riele H. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- 13.Strand M, Earley M, Crouse G, Petes T. Proc Natl Acad Sci USA. 1995;92:10418–10421. doi: 10.1073/pnas.92.22.10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inokuchi K, Ikejima M, Watanabe A, Nakajima E, Orimo H, Nomura T, Shimada T. Biochem Biophys Res Commun. 1995;214:171–179. doi: 10.1006/bbrc.1995.2271. [DOI] [PubMed] [Google Scholar]
- 15.Liu K, Niu L, Linton J P, Crouse G F. Gene. 1994;147:169–177. doi: 10.1016/0378-1119(94)90062-0. [DOI] [PubMed] [Google Scholar]
- 16.Wells J, Held P, Illenye S, Heintz N H. Mol Cell Biol. 1996;16:634–647. doi: 10.1128/mcb.16.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujii H, Shimada T. J Biol Chem. 1989;264:10057–10064. [PubMed] [Google Scholar]
- 18.Drobetsky E, Meuth M. Mol Cell Biol. 1983;3:1882–1885. doi: 10.1128/mcb.3.10.1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caligo M A, Armstrong W, Rossiter B J, Meuth M. Mol Cell Biol. 1990;10:6805–6808. doi: 10.1128/mcb.10.12.6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drummond J T, Genschel J, Wolf E, Modrich P. Proc Natl Acad Sci USA. 1997;94:10144–10149. doi: 10.1073/pnas.94.19.10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marra G, Chang C L, Laghi L, Chauhan D P, Young D, Boland C R. Oncogene. 1996;13:2189–2196. [PubMed] [Google Scholar]
- 22.Hughes M J, Jiricny J. J Biol Chem. 1992;267:23876–23882. [PubMed] [Google Scholar]
- 23.Thomas D C, Roberts J D, Kunkel T A. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 24.Boyer J C, Umar A, Risinger J I, Lipford J R, Kane M, Yin S, Barrett J C, Kolodner R D, Kunkel T A. Cancer Res. 1995;55:6063–6070. [PubMed] [Google Scholar]
- 25.Linton J P, Yen J Y, Selby E, Chen Z, Chinsky J M, Liu K, Kellems R E, Crouse G F. Mol Cell Biol. 1989;9:3058–3072. doi: 10.1128/mcb.9.7.3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Busch D, Greiner C, Rosenfeld K L, Ford R, de Wit J, Hoeijmakers J H, Thompson L H. Mutagenesis. 1994;9:301–306. doi: 10.1093/mutage/9.4.301. [DOI] [PubMed] [Google Scholar]
- 27.Shimada T, Fujii H, Lin H. J Biol Chem. 1989;264:20171–20174. [PubMed] [Google Scholar]
- 28.Kinsella A R, Smith D, Pickard M. Br J Cancer. 1997;75:935–945. doi: 10.1038/bjc.1997.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chabner B A, Allegra C J, Curt G A, Calabresi P. In: The Pharmacological Basis of Therapeutics. Hardman J G, Limbird L E, editors. New York: McGraw–Hill; 1996. pp. 1233–1287. [Google Scholar]
- 30.Gorlick R, Goker E, Trippett T, Waltham M, Banerjee D, Bertino J R. N Engl J Med. 1996;335:1041–1048. doi: 10.1056/NEJM199610033351408. [DOI] [PubMed] [Google Scholar]
- 31.Denis N, Kitzis A, Kruh J, Dautry F, Corcos D. Oncogene. 1991;6:1453–1457. [PubMed] [Google Scholar]
- 32.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1994;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 33.Karran P, Bignami M. BioEssays. 1994;16:833–839. doi: 10.1002/bies.950161110. [DOI] [PubMed] [Google Scholar]
- 34.Anthoney D A, McIlwrath A J, Gallagher W M, Edlin A R, Brown R. Cancer Res. 1996;56:1374–1381. [PubMed] [Google Scholar]
- 35.Jiricny J. Cancer Surv. 1996;28:47–68. [PubMed] [Google Scholar]