Abstract
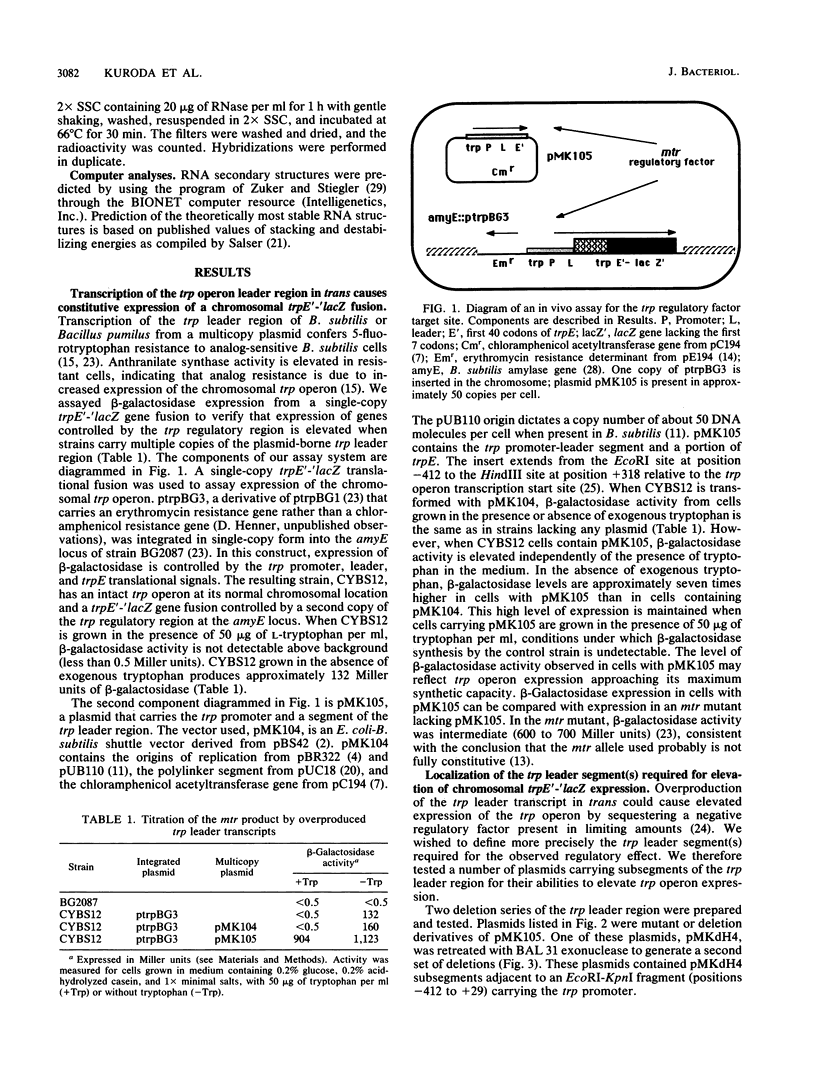
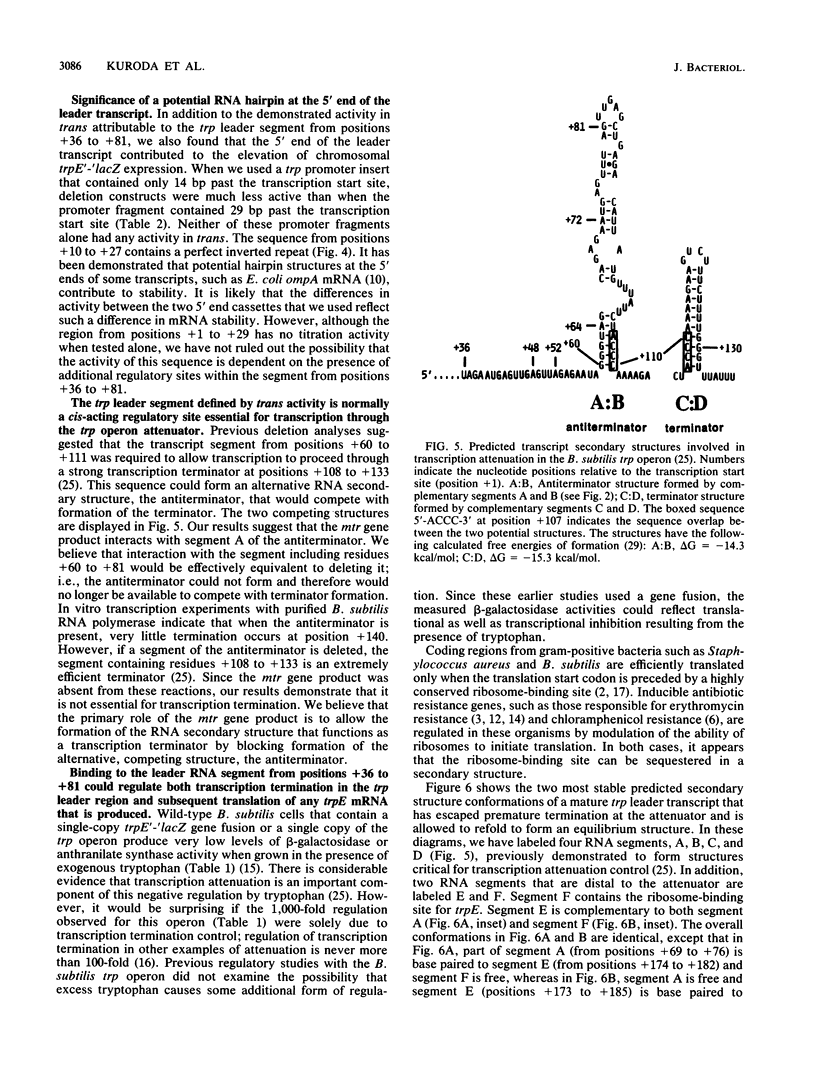
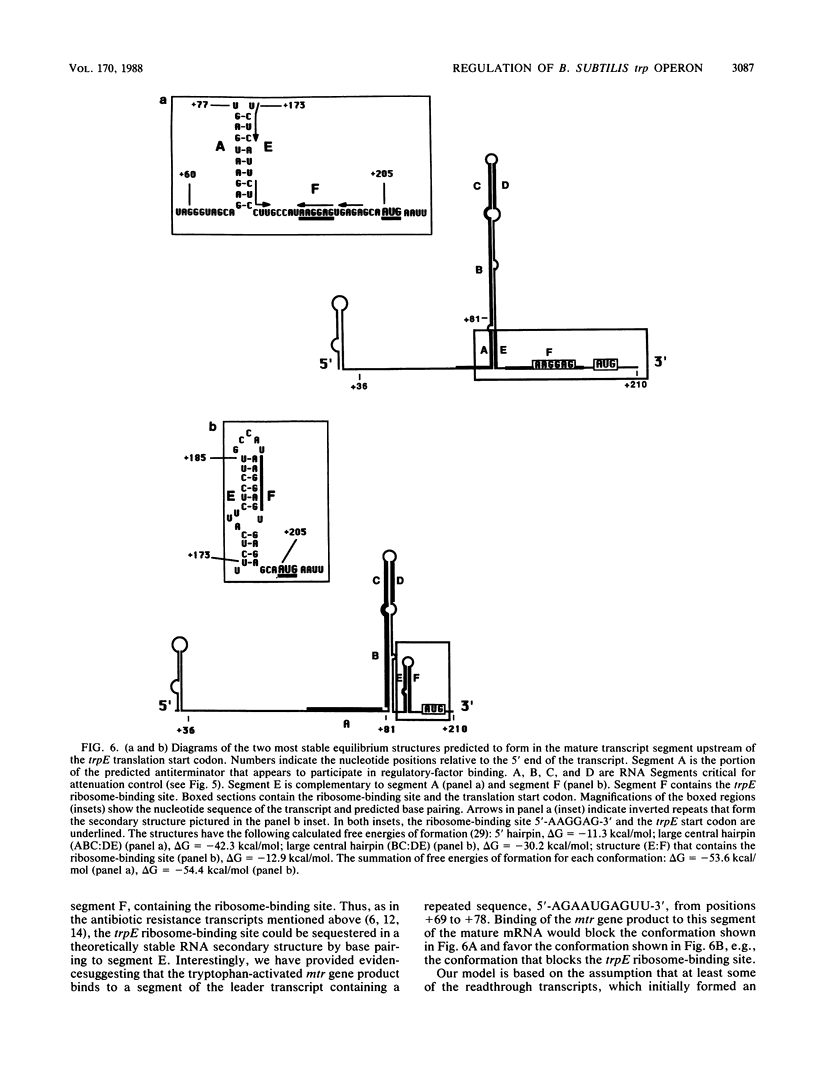
Transcription of the trp operon of Bacillus subtilis is regulated by attenuation. A trpE'-'lacZ gene fusion preceded by the wild-type trp promoter-leader region was used to analyze regulation. Overproduction of the trp leader transcript in trans from a multicopy plasmid caused constitutive expression of the chromosomal trpE'-'lacZ fusion, presumably by titrating a negative regulatory factor encoded by the mtr locus. Subsegments of the trp leader region cloned onto the multicopy plasmid were examined for their abilities to elevate beta-galactosidase activity. An RNA segment spanning the portion of the leader transcript that forms the promoter-proximal strand of the proposed antiterminator structure was most active in this trans test. The data suggest that the mtr gene product, when activated by tryptophan, binds to this RNA segment and prevents formation of the antiterminator. In this manner, the trans-acting factor promotes formation of the RNA structure that causes transcription termination. Secondary-structure predictions for the leader segment of the trp operon transcript suggest that if the mtr factor bound this RNA segment in a nonterminated transcript, the ribosome-binding site for the first structural gene, trpE, could be sequestered in a stable RNA structure. We tested this possibility by comparing transcriptional and translational fusions containing the initial segments of the trp operon. Our findings suggest that the mtr product causes both transcription attenuation and inhibition of translation of trpE mRNA. Inhibition of translation initiation would reduce ribosome density on trpE mRNA, perhaps making it more labile. Consistent with this interpretation, the addition of tryptophan to mtr+ cultures increased the rate of trpE'-'lacZ mRNA decay.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band L., Henner D. J. Bacillus subtilis requires a "stringent" Shine-Dalgarno region for gene expression. DNA. 1984;3(1):17–21. doi: 10.1089/dna.1.1984.3.17. [DOI] [PubMed] [Google Scholar]
- Bechhofer D. H., Dubnau D. Induced mRNA stability in Bacillus subtilis. Proc Natl Acad Sci U S A. 1987 Jan;84(2):498–502. doi: 10.1073/pnas.84.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall E. J., Lovett P. S. Chloramphenicol induces translation of the mRNA for a chloramphenicol-resistance gene in Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3939–3943. doi: 10.1073/pnas.83.11.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich S. D. Replication and expression of plasmids from Staphylococcus aureus in Bacillus subtilis. Proc Natl Acad Sci U S A. 1977 Apr;74(4):1680–1682. doi: 10.1073/pnas.74.4.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Perego M., Hoch J. A. Transcription of Bacillus subtilis subtilisin and expression of subtilisin in sporulation mutants. J Bacteriol. 1988 Jan;170(1):289–295. doi: 10.1128/jb.170.1.289-295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Howard S. M., Hoch J. A. Effect of stage 0 sporulation mutations on subtilisin expression. J Bacteriol. 1986 Apr;166(1):173–179. doi: 10.1128/jb.166.1.173-179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green P. J., Inouye M. Roles of the 5' leader region of the ompA mRNA. J Mol Biol. 1984 Jul 5;176(3):431–442. doi: 10.1016/0022-2836(84)90499-6. [DOI] [PubMed] [Google Scholar]
- Gryczan T. J., Contente S., Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J Bacteriol. 1978 Apr;134(1):318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan T. J., Grandi G., Hahn J., Grandi R., Dubnau D. Conformational alteration of mRNA structure and the posttranscriptional regulation of erythromycin-induced drug resistance. Nucleic Acids Res. 1980 Dec 20;8(24):6081–6097. doi: 10.1093/nar/8.24.6081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch S. O., Roth C. W., Crawford I. P., Nester E. W. Control of tryptophan biosynthesis by the methyltryptophan resistance gene in Bacillus subtilis. J Bacteriol. 1971 Jan;105(1):38–45. doi: 10.1128/jb.105.1.38-45.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Posttranscriptional modification of mRNA conformation: mechanism that regulates erythromycin-induced resistance. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7079–7083. doi: 10.1073/pnas.77.12.7079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M. I., Shimotsu H., Henner D. J., Yanofsky C. Regulatory elements common to the Bacillus pumilus and Bacillus subtilis trp operons. J Bacteriol. 1986 Sep;167(3):792–798. doi: 10.1128/jb.167.3.792-798.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin J. R., Murray C. L., Rabinowitz J. C. Unique features in the ribosome binding site sequence of the gram-positive Staphylococcus aureus beta-lactamase gene. J Biol Chem. 1981 Nov 10;256(21):11283–11291. [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Characterization of the Bacillus subtilis tryptophan promoter region. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6315–6319. doi: 10.1073/pnas.81.20.6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Shimotsu H., Kuroda M. I., Yanofsky C., Henner D. J. Novel form of transcription attenuation regulates expression the Bacillus subtilis tryptophan operon. J Bacteriol. 1986 May;166(2):461–471. doi: 10.1128/jb.166.2.461-471.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizizen J. TRANSFORMATION OF BIOCHEMICALLY DEFICIENT STRAINS OF BACILLUS SUBTILIS BY DEOXYRIBONUCLEATE. Proc Natl Acad Sci U S A. 1958 Oct 15;44(10):1072–1078. doi: 10.1073/pnas.44.10.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmanen I., Lundström K., Lehtovaara P., Sarvas M., Ruohonen M., Palva I. Transcription and translation of foreign genes in Bacillus subtilis by the aid of a secretion vector. J Bacteriol. 1985 Apr;162(1):176–182. doi: 10.1128/jb.162.1.176-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]


