Abstract
Aging is characterized by specific alterations in gene expression, but their underlying mechanisms and functional consequences are not well understood. Here we develop a systematic approach to identify combinatorial cis-regulatory motifs that drive age-dependent gene expression across different tissues and organisms. Integrated analysis of 365 microarrays spanning nine tissue types predicted fourteen motifs as major regulators of age-dependent gene expression in human and mouse. The motif most strongly associated with aging was that of the transcription factor NF-κB. Inducible genetic blockade of NF-κB for 2 wk in the epidermis of chronologically aged mice reverted the tissue characteristics and global gene expression programs to those of young mice. Age-specific NF-κB blockade and orthogonal cell cycle interventions revealed that NF-κB controls cell cycle exit and gene expression signature of aging in parallel but not sequential pathways. These results identify a conserved network of regulatory pathways underlying mammalian aging and show that NF-κB is continually required to enforce many features of aging in a tissue-specific manner.
[Keywords: Aging, NF-κB, epidermis, functional genomics, computational biology]
Genes and environmental factors can influence life span and many phenotypes associated with aging (Mair et al. 2003; Dhahbi et al. 2004; Conboy et al. 2005; Kenyon 2005; Melov et al. 2007). While many mutations can extend life span in model organisms, only physiologic interventions, including caloric restriction (CR), heterochronic parabiosis, and exercise, have been shown to reverse phenotypes of aging in mammals (Mair et al. 2003; Dhahbi et al. 2004; Conboy et al. 2005; Kenyon 2005; Melov et al. 2007). Chronologic aging is associated with diverse and widespread changes in gene expression, which reflect the history and physiologic functions of the aged tissues (Dhahbi et al. 2004; Lu et al. 2004; McCarroll et al. 2004; Fraser et al. 2005). Thus, understanding the control of age-associated transcriptional programs may shed light on mechanisms of aging. However, the large number of genes that become differentially expressed with age and the diversity of such genes between different tissues and species have precluded unifying hypotheses or comprehensive experimental analysis. Here we begin to address this challenge by developing a new analytic approach. We computationally discover combinations of cis-regulatory elements that are predictive of gene expression patterns in individuals of different ages. The regulatory elements then become the common basis of comparison across diverse tissues, physiologic states, and species, and further direct hypothesis-driven experiments.
Using our new bioinformatic approach, we identify the transcription factor (TF) NF-κB as a major regulator of gene expression programs associated with mammalian aging. The NF-κB family of TFs is comprised of five members that each contains a Rel homology DNA-binding domain recognizing the motif GGGRNNYYCC (Hayden and Ghosh 2004; Lim et al. 2007). NF-κB proteins are sequestered in the cytoplasm by binding to IκB proteins; phosphorylation of IκB leads to its degradation and enables nuclear translocation of NF-κB. NF-κB TFs bind DNA as a dimer, the best studied of which is the heterodimeric complex of p50 (encoded by NFKB1) and p65 (encoded by RELA). NF-κB controls the expression of many target genes involved in innate and adaptive immunity, inflammation, and apoptosis; in vitro experiments have suggested that NF-κB may regulate cell senescence (Bernard et al. 2004; Hayden and Ghosh 2004; Hardy et al. 2005; Zhang et al. 2005). Excess NF-κB activity can also induce muscle atrophy (Cai et al. 2004), insulin resistance (Arkan et al. 2005), and neurotoxicity in Alzheimer’s disease (Chen et al. 2005), three prevalent age-associated morbidities. Here we provide evidence for widespread involvement of NF-κB in mammalian aging, and demonstrate a reversal of many features of aging upon acute blockade of NF-κB in aged skin.
Results
Motif module map of aging: genome-scale identification of cis-regulatory motifs driving age-dependent gene expression
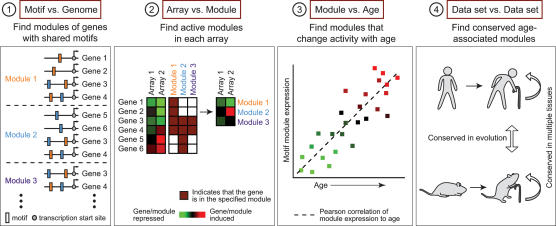
We devised a new analytic pipeline to identify specific cis-regulatory motifs driving age-dependent gene expression programs (Fig. 1). We first assembled groups of genes, termed motif modules, that share upstream motifs for TF binding (see Materials and Methods). We focused on promoter sequences conserved among four mammalian genomes (human, mouse, rat, and dog). Since genes are typically controlled by multiple TFs that act in a combinatorial fashion, we identified modules with all single, pairwise, and three-way combinations of TF motifs in the TRANSFAC database (Matys et al. 2003) and a collection of phylogenetically conserved promoter motifs identified by Xie et al. (2005) (Fig. 1, step 1). To identify the genes that compose each module, we employed a probabilistic model (Sinha et al. 2003) that scores sequences for clustering of TF-binding sites, taking motif degeneracy, frequency, and competition among motifs into account. The algorithm yielded 1095 motif combinations that were significant in a total of 12,680 unique human gene promoters (averaging 425 unique gene promoters per motif combination). Second, in any given gene expression data set, we identified motif modules that were coordinately induced or repressed in each expression profile, thereby detecting specific motif combinations that are activated or deactivated in each experiment (Fig. 1, step 2). Third, we identified specific motif modules that were significantly induced or repressed across samples in an age-associated fashion (P < 0.01, one-sided t-test) (Fig. 1, step 3). Finally, we compared age-dependent motif modules across multiple human and murine tissues to identify candidate age-regulated module that are conserved in each organism and conserved in evolution (Fig. 1, step 4). The end result of the analysis is the prediction of key TF motifs and motif combinations that drive large-scale gene regulation in mammalian aging.
Figure 1.
Motif module map of mammalian aging. (Step 1) Modules of genes that share upstream regulatory sequence motifs are identified. For instance, module 1 consists of all genes with the orange motif, module 2 is the set of all genes with the blue motif, and module 3 is the set of all genes with both orange and blue motifs. (Step 2) The average expression of motif module induction or repression in each microarray gene expression profile is determined; red represents module induction, and green represents module repression. (Step 3) Identification of motif modules that show an age-associated increase (or decrease, not shown) in expression (P < 0.01, one-sided t-test). (Step 4) Conservation of age-associated motifs in multiple tissues and in species identifies candidate general regulators of mammalian aging.
Regulatory motifs involved in mammalian aging
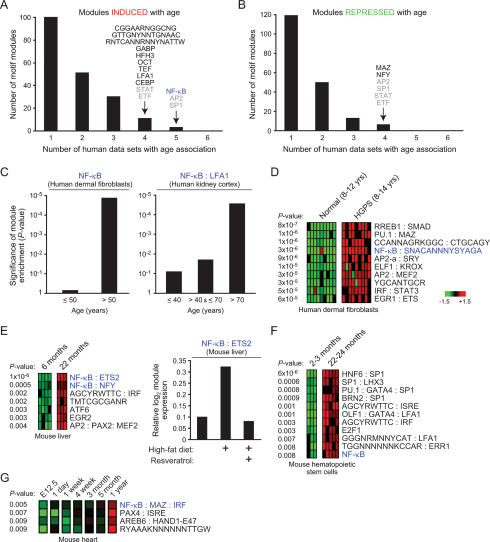
We applied this four-step approach to identify TF motifs that were activated or deactivated with age in 294 microarray profiles representing six human tissue types ranging from 16 to 106 yr of age: skin fibroblasts, kidney cortex, kidney medulla, abdominal muscle, skeletal muscle, and brain (Supplementary Table 1). We hypothesized that motifs representing general regulators of aging may act in multiple tissues, and therefore sought enriched motif that were shared among multiple tissues. While many motifs were induced with age in one of six tissues, only three motifs were induced with age in five of six human tissues, and 11 motifs were induced in four of six tissues (Fig. 2A; Table 1). Four of these motifs (SP1, AP2, STAT, and ETF) in combination with other motifs were also repressed with age in multiple tissues (Fig. 2B), suggesting that these are likely permissive or “noisy” regulatory motifs involved in age-related stochastic changes in gene expression. Notably, three uncharacterized but conserved regulatory motifs show age-dependent induction in human, indicating them as binding sites for candidate regulatory factors involved in aging. A list of all motif module combinations significantly induced and repressed with age in the six human tissues (P < 0.01) is provided in Supplementary Tables 2 and 3, respectively.
Figure 2.
Identification of NF-κB as a regulator of aging-associated gene expression programs. (A,B) Modules induced or repressed with age. Shown is the number of constituent motif modules that are induced (A) or repressed (B) with age in the indicated number of human tissues. Motifs overlapping in four or five human tissues are shown; motifs labeled in gray were found to be both induced and repressed with age in combination with different motifs. NF-κB-containing motif modules are labeled blue throughout the figure. (C) NF-κB module expression increases with chronologic age. Shown is the significance (P-value, hypergeometric distribution) of the fraction of tissue samples within each age group that demonstrates activation of the indicated module. (D) Shown are motif modules significantly induced in Hutchinson Guilford progeria syndrome fibroblasts (P-value, Student’s t-test). Each row is a motif module; each column is a microarray sample. (E, left) Motif module expression in mouse liver. Only modules with P < 0.01 are shown (Student’s t-test). (Right) Relative expression of NF-κB:ETS2 module in mouse liver after the indicated treatments. (F) Motif module expression in mouse haematopoietic stem cells (P < 0.01, Student’s t-test). (G) Motif module expression over time in mouse heart (P < 0.01, one-sided t-test).
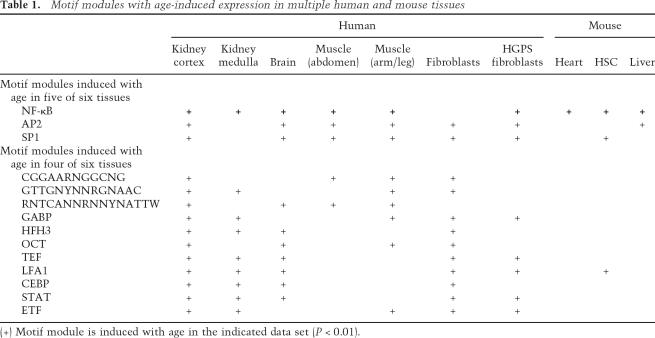
Table 1.
Motif modules with age-induced expression in multiple human and mouse tissues
(+) Motif module is induced with age in the indicated data set (P < 0.01).
We next identified age-induced motifs that showed conservation across human and murine tissues (Table 1; Supplementary Table 1). Intriguingly, the strongest evidence for involvement in age-dependent gene regulation mapped to the binding site of NF-κB TFs, which was a constituent member of motif modules significantly induced with age in nine of 10 human and murine tissues analyzed (Table 1, Fig. 2). The one apparent exception to this trend in normal human skin fibroblasts was due to a technical reason: NF-κB motif module was strongly induced after 50 yr of age but not further induced later, leading to a lower significance based on linear correlation with age (Fig. 2C). In contrast in the remaining human tissues, coordinate induction of modules defined by NF-κB motif alone or by NF-κB in combination with other TF motifs (collectively termed NF-κB motif modules) tended to occur after 40 yr of age and was even more prevalent after 70 yr (Fig. 2C; data not shown). Moreover, a NF-κB motif module was among the top induced modules in cells from Hutchinson Guilford progeria, a disease with features of premature aging (Fig. 2D). Multiple NF-κB motif modules were strongly induced with age in mouse liver, heart, and hematopoietic stem cells (Fig. 2E–G). In an independent data set, treatment of mice with high fat diet induced a NF-κB motif module in the liver, while concomitant treatment with resveratrol, an agent that reverses the effects of high fat diet and prolongs life span (Baur et al. 2006), repressed the module (Fig. 2E). The repeated identification of NF-κB motif modules in this unbiased screen suggests NF-κB as a candidate master regulator of gene expression programs in mammalian aging.
Experimental validation of age-dependent regulation of NF-κB modules
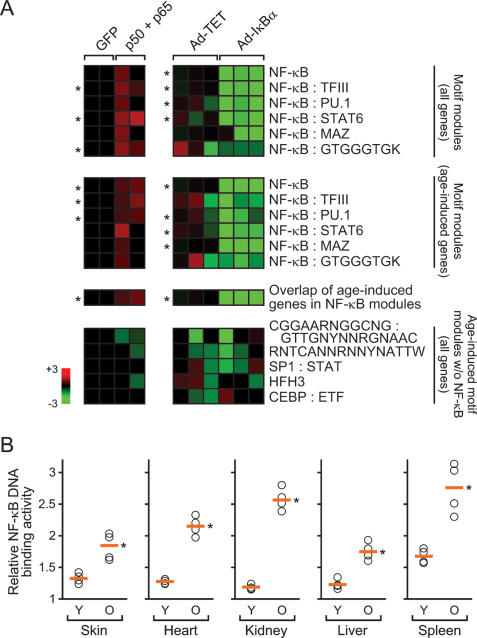
To test that the NF-κB modules we defined computationally indeed represent NF-κB-responsive genes, we verified that six NF-κB-containing motif modules associated with human aging were experimentally responsive to perturbations of NF-κB activity, using published microarray data sets (Fig. 3A; Hinata et al. 2003; H.G. Zhang et al. 2004). Retroviral transduction with NF-κB subunits p50 and p65 significantly induced three of six NF-κB modules compared with control cells transduced with GFP. Conversely, inhibition of NF-κB activity by adenovirus-mediated expression of dominantly active IκBα (Ad-IκBα) significantly inhibited the activation of four of six NF-κB modules in response to TNF-α. The particular fraction of genes within each module that changed with age also showed strong induction upon NF-κB expression and strong repression upon Ad-IκBα expression (Fig. 3A). Importantly, the activities of five additional motif modules that were also induced with age, including two presenting novel motifs, were not altered by NF-κB. This finding supports the specificity of the computationally predicted NF-κB motif modules and suggests that the additional age-associated motif modules may work via pathways that are parallel to and independent of NF-κB. As a second validation, we found that NF-κB DNA-binding activity is induced with age in murine skin, heart, kidney, liver, and spleen (Fig. 3B). These findings are consistent with prior reports on induction of NF-κB DNA-binding activity with age in individual tissues (Chung et al. 2001). We did not observe consistent age-dependent changes in the transcript or protein abundance of NF-κB subunits (Supplementary Fig. 1). The transcripts of genes encoding known regulators of NF-κB activity show diverse age-dependent changes, suggesting that the mechanism of age-dependent NF-κB activation may be tissue specific (Supplementary Fig. 1). Collectively, these results confirm that the computationally defined NF-κB motif modules indeed represent NF-κB responsive genes, and that NF-κB activity is induced with chronologic age.
Figure 3.
Experimental validation of NF-κB motif modules with age. (A) NF-κB-containing motif modules are experimentally responsive to altered NF-κB activity. Each row represents a motif module composed of the indicated genes (the last row represents the overlap of all age-associated genes); each column represents an experiment. Each square is the average expression value of genes in the indicated motif module. NF-κB modules are specifically induced by p50 and p65 expression in primary human keratinocytes (left) and repressed by IκBα expression in primary human fibroblasts (right). (*) P < 0.05 between experimental treatment and control, Student’s t-test. (B) NF-κB DNA-binding activity increase with age in mouse tissue. Shown is the relative level of NF-κB DNA-binding activity in the indicated organ of young (Y; 1 mo) and old (O; 22 mo) wild-type mice. Orange line indicates the average activity. (*) P < 0.005 between young and old for each organ, Student’s t-test.
Inducible genetic blockade reveals a role for NF-κB in enforcement of aging phenotypes
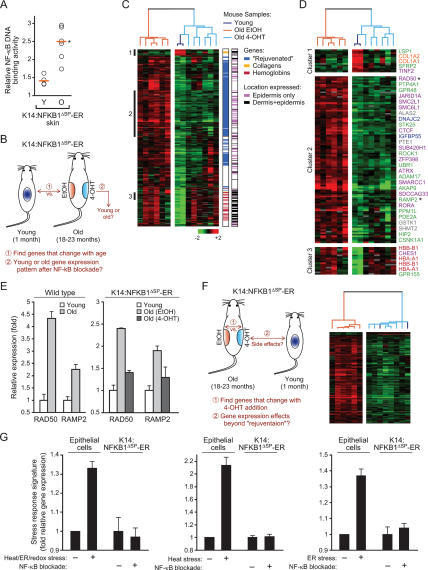
To test the role of NF-κB in mammalian aging, in principle one could examine the life span of mutant animals lacking NF-κB. However, this experiment is not technically possible because NF-κB family members have important developmental functions and RELA−/− mice suffer from embryonic lethality (Hayden and Ghosh 2004). Similarly, although NF-κB activity can be controlled by genetic manipulation of IκB kinases, discovery that the main IκB kinase IKKβ phosphorylates and regulates FOXO3a (Hu et al. 2004), a mammalian homolog of Caenorhabditis elegans aging gene DAF-16, renders interpretation of aging phenotypes in such animals potentially problematic. Instead, we asked whether tissue-specific aging phenotypes might require continuous NF-κB activity in vivo, a question with direct relevance to the treatment of age-related pathologies. We utilized a well-characterized system that allows inducible and site-specific inhibition of NF-κB activity in murine skin, yielding identical phenotypes as epidermal RELA deficiency (J.Y. Zhang et al. 2004; Zhang et al. 2005). The hormone-binding domain of the mutant estrogen receptor (ER) that is responsive to 4-hydroxytamoxifen (4-OHT) was fused to a mutant of p50 (NFKB1ΔSP) that dimerizes with other NF-κB subunits but is unable to bind DNA, thus dominantly inhibiting NF-κB activity (Logeat et al. 1991; Zhang et al. 2005). The fusion protein is normally sequestered in the cytoplasm and inactive; upon 4-OHT addition, NFKB1ΔSP-ER translocates to the nucleus, where it specifically inhibits NF-κB transcriptional activity (Supplementary Fig. 2; Zhang et al. 2005). Experimental analyses confirmed that NFKB1ΔSP-ER is a potent and specific inhibitor of NF-κB DNA-binding activity (mainly the p65:p50 heterodimer) and NF-κB target gene activation (Supplementary Fig. 2). NFKB1ΔSP-ER expression was driven within the basal layer of murine epidermis by the keratin 14 (K14) promoter in transgenic mice. K14:NFKB1ΔSP-ER mice had normal development, life span, and age-dependent induction of NF-κB activity in the skin (Fig. 4A), indicating that NFKB1ΔSP-ER was not constitutively active.
Figure 4.
“Rejuvenation” of age-associated gene expression program upon NF-κB blockade in vivo. (A) NF-κB DNA-binding activity in the skin of K14:NFKB1ΔSP-ER mice increase with age. Orange line indicates the average activity. (*) P = 0.004, Student’s t-test. (B) Schematic of the analysis to identify the fraction of age-associated genes that changes with NF-κB blockade. (C) NF-κB blockade “rejuvenates” age-regulated genes in the epidermis. Expression pattern of 414 genes that changed with age was used to organize all samples (young, old plus EtOH, old plus 4-OHT) by unsupervised hierarchical clustering. (Right) First bar indicates the location of “rejuvenated,” collagen, or hemoglobin genes; second bar indicates the layer-specific expression of each gene in murine skin (April and Barsh 2006). (D) Gene clusters highlighted in C are enlarged. Genes enriched for specific cellular processes are shown (green, protein modification/signal transduction; purple, chromatin/transcriptional regulation; blue, cell cycle/growth control; gray, mitochondrion; orange, collagens; red, hemoglobins). (*) Genes whose expression was verified by qRT–PCR. (E) qRT–PCR validation of age-dependent induction of two aging signature genes in wild-type and K14:NFKB1ΔSP-ER mice. (F, left) Schematic of the analysis to identify expression effects beyond “rejuvenation” upon 4-OHT addition. (Right) NF-κB blockade through 4-OHT addition only induced gene expression patterns seen in young skin. Expression pattern of 276 genes that changed with 4-OHT treatment in old skin was used to organize all samples by unsupervised hierarchical clustering. Note that the two young samples are intermixed with old 4-OHT-treated samples in the dendrogram. (G) NF-κB blockade in old skin does not induce transcriptional signatures of cell stress. Shown is the average relative gene expression (±SD) of all genes that comprise the transcriptional signatures in response to heat, ER (tunicamycin or DTT), or redox (menadione) stresses (left); all genes in response to heat alone (middle); or all genes in response to ER stress alone (right) in epithelial cells (Murray et al. 2004) and the average expression of these genes in old K14:NFKB1ΔSP-ER mice with and without NF-κB blockade.
To test whether NF-κB activity is required to maintain age-associated gene expression, we applied topical 4-OHT and ethanol vehicle (EtOH) for 2 wk to the right and left dorsal flanks, respectively, of chronologically aged transgenic mice (18–23 mo) (Fig. 4B). The use of spatially controlled NF-κB blockade allowed comparison with the contralateral, aged tissue on the same mouse, thereby controlling for many stochastic and unknown factors that may arise in different aged animals. Young transgenic mice (1 mo) were treated in parallel as control. Skin samples were harvested and analyzed for gene expression using mouse cDNA microarrays containing ∼19,000 unique genes. As expected, canonical NF-κB target genes were coordinately repressed by 4-OHT treatment in old transgenic mice but not in wild-type mice (Supplementary Fig. 3). To rigorously define aging in molecular terms, we first identified all genes whose expression changed with age in this system, and then we tested how NF-κB blockade altered the expression of these age-associated genes (Fig. 4B). Comparison of global expression profiles of young skin samples with the EtOH-treated old skin revealed 414 genes that were significantly altered (mostly induced) in old skin (false discovery rate [FDR] < 0.05 using algorithm Significance Analysis of Microarrays [SAM] [Tusher et al. 2001]) (genes listed in Supplementary Table 4). This set of murine age-associated genes is enriched for the presence of NF-κB-binding sites in their promoters (60%, P < 0.01; see Materials and Methods), experimentally confirming the evolutionarily conserved role of NF-κB in aging. Upon NF-κB blockade in old skin, expression of 225 of these 414 genes (54%) was reduced to expression levels indistinguishable from those of the young samples (Fig. 4C,D). Unsupervised hierarchical clustering confirmed that the expression pattern of 4-OHT-treated skin was globally more similar to that of young skin than the contralateral EtOH-treated skin on the same aged animals (Fig. 4C). These results reveal that NF-κB activity is required to maintain a substantial portion of the global gene expression program induced with age in murine skin in vivo.
Age-associated genes whose expression was inhibited by NF-κB blockade are multifaceted and notably included those related to chromatin/transcriptional regulation (RAD50, SMC2L1, SMC6L1, and ATRX), protein modification/signal transduction (STK25, RAMP2, and HIP2), cell cycle/growth control (DNAJC2 and IGFBP5), and mitochondrion (ALAS2, GSTK1, and PTE1) (Fig. 4D, cluster 2; Supplementary Fig. 4). Genes related to inflammation and immunity were not substantially affected by NF-κB blockade in this context, nor did we see evidence of inflammatory infiltrate with aging (data not shown). Quantitative RT–PCR (qRT–PCR) analysis confirmed the age-dependent induction and reversal by NF-κB blockade of RAD50 and RAMP2, two genes tested from the signature; we also confirmed that these same genes are induced by chronologic age in wild-type mouse skin (Fig. 4E).
Because K14:NFKB1ΔSP-ER is expressed only in the epidermis, the incomplete “rejuvenation” of the gene expression program of whole skin by NF-κB blockade may be due to the presence of other cell types that do not express the transgene. Indeed, two main clusters of age-associated genes in skin that were not altered by NF-κB blockade were enriched for collagens (COL1A1, COL1A2) expressed predominantly by dermal fibroblasts (Fig. 4D, cluster 1) and adult hemoglobins (HBA-A1, HBB-B1) likely expressed by reticulocytes within the vasculature (Fig. 4D, cluster 3). Further, systematic comparison of our data with the layer-specific transcriptional profiles of murine skin (April and Barsh 2006) revealed that epidermal-specific expression strongly predicted the “rejuvenation” of age-related genes’ expression pattern by epidermal NF-κB blockade (Fig. 4C, Supplementary Table 4). Eighty-five percent of age-associated genes that were “rejuvenated” by epidermal NF-κB blockade had restricted expression in the basal or suprabasal layers of epidermis, while 83% of the age-associated genes that failed to be “rejuvenated” by NF-κB blockade had expression in the dermis (P < 10−11, χ2 test). The density of predicted NF-κB-binding sites was otherwise not different among promoters of “rejuvenated” and unresponsive age-induced genes. Together, these results suggest that the observed “rejuvenation” of gene expression occurs mostly through blockade of age-related genes in the epidermis.
Next, we tested whether NF-κB blockade by 4-OHT treatment induced transcriptional changes other than reversal of aging-associated gene expression (Fig. 4F). Genes whose expression differed between EtOH- and 4-OHT-treated old samples were identified using SAM. The expression levels of these genes in the young samples were then observed through unsupervised hierarchical clustering of all samples. Changes induced by inhibiting NF-κB were very specific because all of the expression changes induced by 4-OHT showed a similar expression pattern as the young samples (Fig. 4F). No gene was significantly altered by 4-OHT treatment that was not also reciprocally altered by aging (FDR < 0.05 cutoff). Control experiments with nontransgenic mice confirmed that 4-OHT alone did not cause “rejuvenation” of gene expression (Supplementary Fig. 5). NF-κB blockade also did not induce either apoptosis or gene expression signatures of cell stress (Murray et al. 2004), including canonical transcriptional responses to oxidative, protein unfolding, or heat stresses (Fig. 4G, Supplementary Fig. 6). Thus, the gene expression differences we observe are due to NF-κB inhibition and act specifically to revert epidermal gene expression to that observed in young skin.
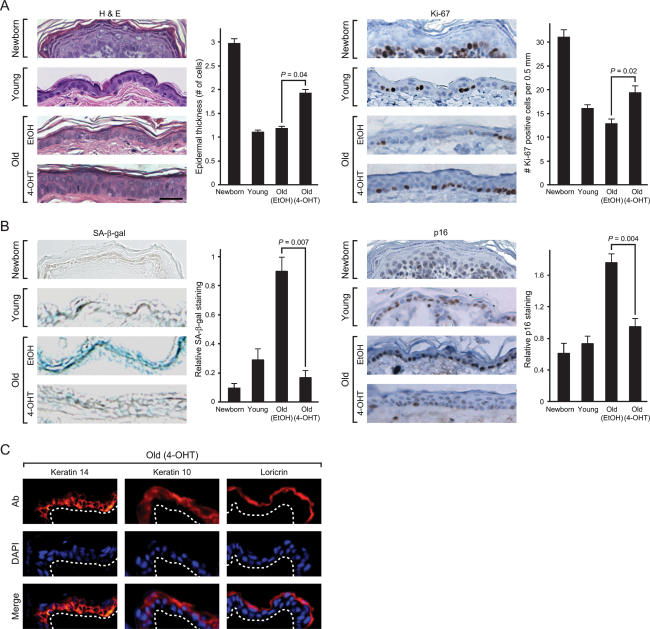
In addition to alterations in gene expression, aged skin is characterized by epidermal atrophy, decreased proliferative capacity, and increased frequency of cellular senescence (a state of irreversible cell cycle arrest). Newborn murine epidermis is three to five cell layers thick, but typically becomes one to two cells thick after 1 mo of age. Aged murine tissues also exhibit increased expression of SA-β-gal (Dimri et al. 1995) and p16INK4A (Krishnamurthy et al. 2004), two markers of cellular senescence. While the epidermis of 1-mo-old and aged EtOH-treated skin samples was one cell thick, 4-OHT treatment increased epidermal thickness in old skin to a degree intermediate between newborn and 1-mo-old skin (Fig. 5A, left). Consistent with the increase in epidermal thickness, NF-κB blockade in aged epidermis increased cell proliferation, as indicated by increased expression of the proliferation marker Ki-67 (Fig. 5A, right). As expected, SA-β-gal activity and p16INK4A protein expression were induced in aged skin, but both markers of senescence were significantly decreased upon NF-κB blockade (Fig. 5B). Six additional gene markers of senescence, identified through microarray analysis following oncogene-induced senescence (Hardy et al. 2005), were also found to be induced with age and strongly resilenced upon NF-κB blockade (Supplementary Fig. 7). Importantly, reversal of cell senescence and increased proliferative capacity induced by NF-κB blockade occurred with preservation of normal tissue homeostasis and differentiation. In stratified epithelium, proliferative basal cells adherent to the underlying basement membrane undergo cell cycle arrest then outward migration and terminal differentiation. Cell proliferation after NF-κB blockade occurred predominantly in the basal layer of epidermis, the normal proliferative compartment (Fig. 5A), and the spatial organization of the mature epidermal stratification program, including K14 (basal), K10 (suprabasal), and loricrin (granular layer), was intact (Fig. 5C). K6 and K17, markers of hyperproliferation and truncated epidermal differentiation, were not induced (Supplementary Fig. 8). We also did not observe induction of K8 and K18, markers of simple epithelia, or defects in hair follicle shape and function, which are indicative of reversion to embryonic stages of skin development (data not shown). Together, these results suggest that NF-κB activity is continually required to maintain cellular senescence associated with chronologic aging in murine skin.
Figure 5.
Reversal of tissue characteristics of epidermal aging by NF-κB blockade. (A) NF-κB inhibition increases epidermal thickness and proliferation. (Left) Hematoxylin and eosin (H&E) staining of mouse epidermis. Average epidermal thickness (number of cell layers) is shown (±SEM). P-values show significant difference between old EtOH and old 4-OHT (Student’s t-test). Bar, 30 μm. (Right) Immunohistochemistry of Ki-67 protein expression in the epidermis of paraffin-embedded sections is shown (average number of Ki-67-positive cells ± SEM). (B) NF-κB blockade decreases senescence markers SA-β-gal and p16INK4A. (Left) SA-β-gal activity in the epidermis of frozen sections is shown (average SA-β-gal activity on a 0–3 scale ± SEM). (Right) Immunohistochemistry of p16INK4A protein in paraffin-embedded sections is shown (average p16INK4A staining on a 0–3 scale ± SEM). (C) Immunofluorescence of K14, K10, and loricrin (Ab, red) in frozen sections of old skin after NF-κB blockade. Nuclei are blue with DAPI counterstain. White dashed line indicates basement membrane.
NF-κB regulation of cell cycle exit and gene expression signature of aging is separable
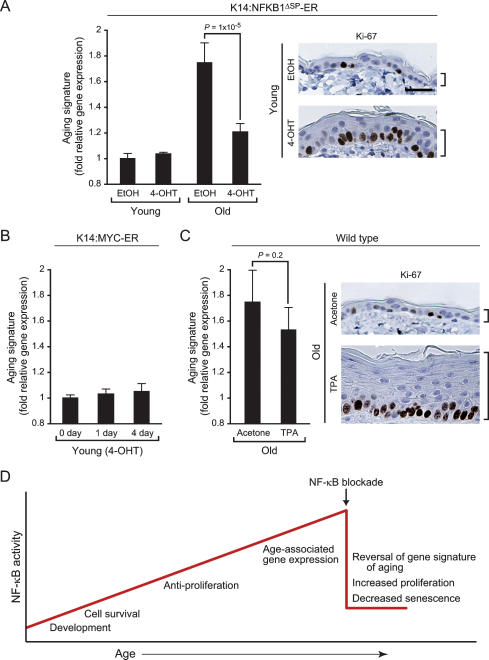
The ability of NF-κB blockade to revert the global gene expression program and tissue characteristics to those more resembling younger mice raises the question whether these two events are mechanistically linked. NF-κB has been implicated in the induction of cell senescence and cell cycle exit; therefore, one possibility is that the gene expression signature of epidermal aging arose as a consequence of cell cycle exit imposed by NF-κB. Alternatively, cell cycle exit and gene expression signature of aging may be independent functions of NF-κB. To distinguish between these two possibilities, we first compared age-specific effects of NF-κB blockade in the epidermis (Fig. 6A). NF-κB blockade in young skin (by 4-OHT treatment of 4-wk-old K14:NFKB1ΔSP-ER mice) led to increased cell proliferation but did not alter the expression of the gene expression signature of aging, indicating that the consequence of NF-κB action is age specific and separable from cell cycle exit. Consistent with this observation, enforced cell proliferation in young K14:MYC-ER transgenic mice induced epidermal cell proliferation and expression of hyperproliferative keratins (K6 and K17) with no effect on the aging gene expression signature (Fig. 6B, Supplementary Fig. 8). To further demonstrate the specific role of NF-κB in the transcriptional program of aging, we analyzed the genomic expression programs of skin or primary keratinocytes in response to the genetic activation or deletion of four additional TFs (cJun, β-Catenin, p63, and IRF6). While each of these factors has a demonstrable phenotype, none of them affected the gene signature of aging (Supplementary Fig. 8). Finally, we induced epidermal proliferation in old wild-type mice and asked if enforced cell cycle exit could suppress the gene expression signature of aging. Treatment of skin in old wild-type mice with the drug TPA induced massive epidermal proliferation, but microarray analysis showed that TPA had little effect on the aging gene expression signature (Fig. 6C). Together, these data suggest that anti-proliferation and age-dependent gene expression are two separable functions of NF-κB. The former is operative in both young and old tissue with modest level of NF-κB while the latter is a context-specific and novel function of accumulated NF-κB in physiologically aged tissues (Fig. 6D).
Figure 6.
NF-κB controls cell cycle exit and gene expression signature of aging in parallel but not in sequential pathways. (A) NF-κB inhibition in young mice induces epidermal proliferation without affecting gene expression signature of aging. (Left) Average relative gene expression (±SD) of the aging signature (genes that are induced by age and reversed by NF-κB blockade from Fig. 4C) in young and old K14:NFKB1ΔSP-ER mice. P-value shows significant difference between old EtOH and old 4-OHT (Student’s t-test). (Right) Ki-67 protein expression and epidermal thickness before and after 4-OHT treatment. Bar, 30 μm. (B) Enforced cell cycle entry by MYC does not induce the aging signature. Shown is the average relative gene expression (±SD) of the aging signature 1 or 4 d after 4-OHT treatment in young K14:MYC-ER mice. (C) TPA-enforced hyperproliferation in old skin has little effect on the aging signature. Gene expression and Ki67 staining are as above. (D) Model of NF-κB action in aging. Accumulation of NF-κB activity with age imparts additional layers of age-specific functions. Upon NF-κB blockade in aged tissue, multiple features characteristic of aging are reversed.
Discussion
Motif module map of aging as an engine of biological discovery
We developed a pipeline for systematic analysis of cis-regulatory motifs in aging that has several advantages. First, microarray analysis of aging (and of many other biological states) is complicated by the potentially large list of subtly differentially expressed genes, which precludes experimental analysis in a comprehensive manner. Our approach seeks to identify upstream regulators of the large-scale transcriptional changes, which enables investigators to manipulate the key pathways impinging on the motifs and thus experimentally manipulate the gene signature of interest to identify its functional consequences. Second, consideration of gene regulation at the level of modules is substantially more powerful than traditional gene-level methods (Segal et al. 2005; Subramanian et al. 2005). The collaboration of a cis-regulatory motif with different tissue-specific motifs provides a possible explanation for how a conserved mechanism may give rise to diverse age-associated changes in gene expression among different tissues (McCarroll et al. 2004; Fraser et al. 2005). Such a mechanism can be missed by gene-level analyses seeking overlap in gene expression changes among different aged tissues (McCarroll et al. 2004) but is detected by our motif module method that captures the combinatorial and conditional activities of higher level gene modules. Third, with the rapid accumulation of gene expression data in a variety of systems, our method provides an unbiased and scalable strategy to integrate and leverage diverse models of aging. For instance, we identified 11 known and three uncharacterized motifs that are induced with age across diverse human and mouse tissues, diseases, and age-related experiments. Detailed in vitro and in vivo analysis of NF-κB, a TF family nominated by the motif module map, confirmed its age-specific and ongoing role to enforce the global transcriptional program and tissue phenotypes of epidermal aging. These results highlight the potential utility of our computational methods for genomic studies of aging and potentially other complex physiologic processes.
Reversibility of mammalian aging phenotypes
Aging is thought to reflect the accumulation of stochastic cellular damages and the effects of key genetic programs that act throughout life to control longevity (Kenyon 2005). CR and heterochronic parabiosis can restore many features of youth to aged mammalian tissues (Mair et al. 2003; Dhahbi et al. 2004; Conboy et al. 2005), and expression of telomerase can reverse the effects of replicative senescence in vitro (Funk et al. 2000), suggesting that some features of mammalian aging may be plastic. Our data suggest that many molecular and cellular features associated with mammalian aging, at least in epidermis and possibly other tissues, may be actively enforced until late in life, and age-associated phenotypes can be substantially reversed by a single gene intervention. Furthermore, NF-κB action in skin aging appears to be cell-autonomous since we were able to reverse multiple characteristics of aging in patches of epidermis by NF-κB blockade in otherwise old animals. These results bode well for the possibility of targeted therapies to reverse features of aging to alleviate age-related pathologies in the elderly. An important caveat of our study is the transience of NF-κB blockade; optimal strategies of NF-κB blockade to induce healthful effects in aged tissues in the long term need to be addressed in future studies.
NF-κB enforcement of aging
As a TF that is responsive to oxidative stress, DNA damage, growth signals, and immune activation (Hayden and Ghosh 2004), NF-κB is centrally placed to transduce diverse extracellular signals to adaptive changes in gene expression and tissue homeostasis (Chung et al. 2001). Indeed, several additional regulators of aging functionally interact with NF-κB. SIRT1, the mammalian homolog of yeast SIR2 gene that mediates replicative and CR-extended life span, binds to and deacetylates p65 RELA, thereby inhibiting NF-κB activity (Yeung et al. 2004). FOXO3a, a homolog of the C. elegans longevity gene DAF-16, represses NF-κB nuclear translocation and transcriptional activity (Lin et al. 2004). Intriguingly, the human NFKB1 gene maps within a genetic locus on chromosome four that has been repeatedly associated with human longevity (Reed et al. 2004). Recent studies in model organisms have suggested that target hub proteins that receive input from multiple upstream pathways are likely key integration and control points in morphogenetic pathways (Borneman et al. 2006). Application of similar criteria to the network architecture of genes involved in aging may help to prioritize additional regulatory motifs that may play dominant roles in mammalian aging.
Our study highlights several areas for future studies. First, the mechanism by which NF-κB becomes activated with age remains unclear. Examination of promoter occupancy of NF-κB target genes in vivo in young and old tissues may be useful to elucidate the transcriptional complexes enforcing gene activation. Second, the contribution of specific NF-κB target genes to aging may be examined. Interestingly, seven NF-κB target genes identified in this study possess homologs that extended life span when inactivated in C. elegans (Supplementary Table 5). It should also be noted that given the large number of NF-κB responsive genes (hence our emphasis on gene modules instead of single genes), the biological effects of NF-κB blockade may not be mediated by just a small number of target genes. For instance, the C. elegans DAF-16 gene encodes a forkhead TF that mediates longevity by controlling the expression of hundreds of genes. Systemic RNA interference (RNAi) studies of a large number of DAF-16-dependent genes showed that individual target genes had at best very modest influence on aging. It was the combined effect of all the target genes that explained the potency of DAF-16 (Murphy et al. 2003). In summary, our identification of age-dependent NF-κB motif modules and candidate target genes provide a framework for systematic dissection of transcriptional programs of aging in the future.
Materials and methods
Gene expression data sets of human aging
Microarray analysis of primary human fibroblasts from individuals of different ages is described below. We selected five other published microarray data sets of aging in human tissue for comparison (Supplementary Table 1). Our criteria were (1) the full data set (not just selected gene lists) be available for download, as our method required genome-wide measurements, and (2) the data set contained large numbers of samples of individuals of a wide range of ages to ensure greater power for statistical analysis. We used EntrezGene ID (previously LocusLink ID) to map probes among different microarray platforms.
Culture and microarray procedures for aged skin fibroblasts
We profiled 49 primary cultures of dermal fibroblasts from normal, healthy individuals of ages 17–92 yr, collected in the National Institute of Aging’s Baltimore Longitudinal Study of Aging (Coriell Cell Repository). We selected fibroblasts that (1) represented a broad span of age groups equally, (2) had ≤10 passages, and (3) had no chromosomal abnormalities as determined by FISH. Fibroblasts were propagated in an identical manner in the absence of connective tissue using Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Hyclone), glutamine, and 100 U penicillin–streptomycin (GIBCO). Cells were passaged for five to 10 population doublings in vitro before mRNA harvest. Construction of human cDNA microarrays, mRNA purification using FastTrack (Invitrogen), and array hybridizations were performed as described previously (Chang et al. 2002).
Motif module map of aging
Each motif module is a group of genes that have in common the presence of one or more specific TF-binding motifs in their upstream regulatory regions (Fig. 1, step 1). We defined the upstream regulatory region of each gene as in Xie et al. (2005). to be the (up to) 4000 base pairs (bp) centered at the annotated transcription start site and conserved among human, mouse, dog, and rat genomes. Redundancies were detected using BLAST (score, >500 bits) and removed to give a total of 13,350 human sequences. We downloaded DNA-binding sites from TRANSFAC (Matys et al. 2003) and Xie et al. (2005) and represented each binding site as a position-specific scoring matrix (PSSM). For all single, pairwise, and three-way combinations of these motifs, we then identified the set of genes that significantly contain the particular combination in their regulatory region. To this end, for each motif combination and regulatory region of a particular gene, we used a probabilistic model (Sinha et al. 2003) to assign a score that represents the overall likelihood that the regulatory region contains the combination of motifs (normalized against background probability inferred from the local nucleotide composition of the same sequence). The probabilistic model treats the PSSMs as prescribing binding free energies and takes frequency and competition among PSSMs into account when computing the score of a particular regulatory region. When identifying pairwise and three-way combinations of motifs, we excluded redundant combinations (as the motif databases used have sets of highly similar motifs), by only considering higher-order combinations whose score improves compared with the score of each combination of motifs from the previous level. A single motif is considered as “targeting” a regulatory region if its score has an empirical P-value <0.01, estimated from 100 random permutations of that region. Specifically, we quantify the distribution of scores that would be expected by chance if the base composition of the promoter is held constant and if the promoter is assumed to not include the binding site motifs. A motif combination’s target set is the intersection of the target sets of its component motifs; the P-value of the score for motif combinations uses a model that has the combination of motifs. We rejected motif combinations whose target sizes were not significantly large (FDR < 0.05, hypergeometric distribution). Mouse motif modules were constructed by mapping reciprocal best BLAST hits between human and mouse genes and assigning the same module memberships to each mouse ortholog.
After identifying motif modules based on genomic sequences, the next task is to determine which motif module(s) can potentially explain the observed gene expression data (Fig. 1, step 2). For each microarray profile, we identified the extent of module induction or repression by averaging the expression of all member genes and separating them into induced modules (expression change, >0 in log2 space) or repressed modules (expression change, <0 in log2 space). The average expression of the induced or repressed genes is obtained for every motif module in every microarray profile. This analysis yields a higher-level view of each expression profile as a combination of activated and deactivated motif modules.
Next, we identified motif modules that tended to be induced or repressed in an age-associated fashion (Fig. 1, step 3). We calculated the Pearson correlation of the average expression levels of each module to the age of the samples. Positive Pearson correlation values were obtained for modules whose expression level increased with age; negative Pearson correlation values were obtained for modules whose expression level decreased with age. One-sided t-tests (calculated with Winstat, R. Fitch Software) were used to determine the significance of each Pearson correlation, and motif modules were classified as induced or repressed with chronologic age if P-value was <0.01 (see Supplementary Tables 2, 3 for complete list).
Last, we searched for motifs that were regulated with age in multiple tissues and organisms (Fig. 1, step 4). We first examined all of the enriched motif modules and their component motifs in six human data sets. Fourteen constituent motifs were identified as induced with age in four or five of the data sets; six motifs were found to be repressed with age in four of the six data sets analyzed. We next analyzed the expression of the 14 age-induced motifs in four additional human and mouse data sets. One mouse expression data set was a time course and was analyzed as above (Fig. 1, step 3) for the human data sets (motifs with P < 0.01 were isolated). Each of the remaining human and mouse data sets contained replicates of young or old tissue samples; the expression of each motif module was obtained as above (Fig. 1, step 2) in each sample, and Student’s t-tests were used between the young and old groups to identify motifs with P < 0.01.
Because motif occurrence in the genome is variable, we controlled for false discovery due to frequent motif occurrence by several means. First, the motif module map aimed to provide a comprehensive and accurate inventory of motif frequencies by comparing the occurrence of motif-matching sequences versus the permuted sequences of the same promoter regions. Thus, nucleotide content (which can vary widely over the genome) is preserved in our comparison. Second, we take motif frequency into account when we parse gene expression profiles of aging by our motif modules, by considering the average expression level of all genes in each particular motif module. Thus, for a large motif module comprised of many member genes, a correspondingly larger number of genes must be coordinately induced or repressed to influence the module average compared with a small module comprised of fewer genes. For instance, one of the largest motif modules we recovered was one that corresponded to CpG islands, which is present in ∼30% of mammalian promoters. We did not find any evidence for this module to be coordinately induced or repressed with age in any of the data sets examined (data not shown).
Mouse studies
K14:NFKB1ΔSP-ER vector and the K14:NFKB1ΔSP-ER transgenic line were as described (Zhang et al. 2005). The K14:MYC-ER line was as described (Frye et al. 2003). Old wild-type mice (C57BL/6) were obtained from the National Institute on Aging. NF-κB blockade in young and old K14:NFKB1ΔSP-ER mice was induced by topical application of 4-OHT (1–1.5 mg per day in EtOH) or EtOH as a control on dorsal skin for 2 wk. Hyperproliferation in old wild-type mice was induced by topical application of 12-O-Tetradecanoylphorbol-13-acetate (TPA; 6.5 μg every other day in acetone) or acetone as a control on dorsal skin for 2 wk.
Electrophoretic mobility shift assays (EMSA) and immunoprecipitation (IP) assays
NF-κB DNA-binding activity in mouse skin was determined by NoShift EMSA (Novagen) per the manufacturer’s instructions. NF-κB DNA-binding activity and supershift assays in retrovirally transduced primary human keratinocytes were analyzed using Promega Gel Shift Assay System; γ-32P was obtained from PerkinElmer. For IP of NFKB1ΔSP-ER in transduced primary human keratinocytes, cell lysis, IP, and wash steps were performed in IP buffer (50 mM Tris-HCl at pH 7.4, 250 mM NaCl, 0.25% Triton X-100, 10% glycerol). Following overnight incubation of cell lysates with anti-ERα (MC-20; Santa Cruz Biotechnology), antibody–protein complexes were purified with immobilized Protein A/G (Pierce) and eluted by boiling. The following antibodies for EMSA supershift and IP assays were from Santa Cruz Biotechnology: anti-p65 (H-286), anti-p50 (H-119 or C-19), anti-cRel (C), anti-p52 (K-27 or C-5), and anti-RelB (C-19).
Microarray analysis of murine skin
Total RNA was extracted with TRIzol (Invitrogen), and RNA was amplified using the Ambion MessageAmpII aRNA kit. Amplified total Mouse Universal Reference RNA (Stratagene) was used as reference. cDNA microarray construction and hybridization were as described (Miki et al. 2001).
Data analysis of NF-κB blockade in vivo
For analysis, we selected genes in which the corresponding array element had fluorescent hybridization signal at least 1.5-fold over local background in either Cy5 or Cy3 channel, and also had technically adequate data in at least 50% of experiments. Expression value of each gene was compared with its mean value across all experiments.
Effect of NF-κB blockade on age-associated gene expression program
We identified all genes that were significantly regulated with age, and then tested how NF-κB blockade in old mice affected the expression of these genes. To identify gene expression that changed with age, we carried out a two-class comparison using the permutation-based algorithm SAM (Tusher et al. 2001) between young samples and EtOH-treated old samples (FDR < 0.05). After this initial selection, we noticed that a small number of genes made the cutoff by virtue of being consistently repressed in young samples but were variably expressed in old samples. They were strongly induced in some old samples but repressed others; therefore, their average fold of induction across all old samples was near zero. We deemed these to be “noisy” genes that should be removed in order to preserve a more robust and consistent list of genes as biomarkers of aging. Using mean-centered data, we calculated the average level of expression in old versus young samples and removed genes whose mean level of expression did not change >0.1-fold in log2 space (i.e., genes whose average fold induction across all old samples was near zero); 414 probes (henceforth, genes) met these criteria for age-associated changes and are listed in Supplementary Table 4. Next, we tested how NF-κB blockade in old samples affected the expression of these 414 genes. We defined “rejuvenated” genes as genes whose expression pattern was significantly different between 4-OHT- and EtOH-treated old samples and became more similar to that of young samples. Two-class comparison of 4-OHT- and EtOH-treated old samples by SAM (FDR < 0.07) and further comparison with young samples yielded 225 of 414 genes (54%) that met these criteria. To determine the effect of NF-κB blockade on the aging signature in the skin of young mice, we averaged the expression of all 225 “rejuvenated” genes following EtOH or 4-OHT treatment of young K14:NFKB1ΔSP-ER skin and compared this with the average expression of the same genes in old K14:NFKB1ΔSP-ER skin. qRT–PCR to validate two of the “rejuvenated” genes was carried out on total RNA using the TaqMan quantitative one-step RT–PCR as described by the manufacturer (Applied Biosystems), normalized to GAPDH levels.
Identification of NF-κB-binding sites in age-associated genes
For each age-associated gene, the 10,000 bp upstream of the annotated transcription start site in the mouse genome was analyzed in 500-bp windows for NF-κB-binding sites. For each window, we analyzed the presence of position-weighted matrices of NF-κB-binding sites and calculated the P-value as the frequency of NF-κB-binding sites in 1000 order-permuted sequences of the window length versus the observed NF-κB-binding sites in the genomic sequence. Of the 414 age-associated genes, 216 had unique LocusLink IDs; 174 of these genes had annotated transcription start sites and sequence information. Motif module map was used to identify NF-κB-binding sites in 105 of these genes (60%, P < 0.01) (Supplementary Table 4).
Side effects of NF-κB blockade
We identified all genes that changed with NF-κB blockade in old skin and then tested whether any of the induced transcriptional patterns were not observed in young skin. To identify genes that changed upon 4-OHT addition, we carried out a two-class comparison with SAM between old EtOH- and 4-OHT-treated samples, obtaining 276 genes (FDR < 0.05). Unsupervised hierarchical clustering of these 276 genes across all old and young samples revealed that the young samples were intermixed with 4-OHT-treated old samples, indicating that NF-κB blockade induced an expression pattern that was not distinct from young skin.
4-OHT effect in nontransgenic mouse
To control for gene expression changes induced by the drug 4-OHT, we carried out the same drug/control treatment as described above in duplicate on a 20-mo-old nontransgenic mouse that was of identical genetic background. SAM was used to identify genes that changed expression between EtOH- and 4-OHT-treated samples. No genes were significantly induced or repressed upon 4-OHT addition (FDR > 0.19).
Layer-specific transcriptional profiles in skin of age-associated genes
Using recently published skin layer-specific transcriptional profiles (April and Barsh 2006), we identified the skin layer in which each age-associated gene is normally expressed. We intersected the data sets using Unigene identifier and were able to determine the expression location for 166 of 414 age-associated genes (Supplementary Table 4). Sixty-seven of the 94 epidermal-specific genes were “rejuvenated,” while only 12 of the 72 genes expressed in the dermis and epidermis were “rejuvenated”; thus, epidermal-specific genes are highly enriched for “rejuvenated” genes (P < 10−11, two-by-two χ2 test).
Gene expression signatures of cell stress after NF-κB blockade
To test whether NF-κB blockade induced gene expression signatures of cell stress, we intersected our expression data with that of expression data of cultured epithelial cells in response to various stresses (Murray et al. 2004). We mapped human–mouse gene orthologs by reciprocal best hit using Blast. Three signatures of genes that showed a consistent response to stress in HeLa cells were isolated: (1) general response to multiple stresses, (2) heat-shock response, and (3) response to 2.5 mM of dithiothreitol (DTT), an ER stress. The stress response, as measured by the average expression of all genes in each stress cluster at three time points, was determined for the same genes in 4-OHT-treated mice compared with EtOH-treated mice. We did not observe any induction of these stress gene expression signatures upon NF-κB blockade (P > 0.1, Student’s t-test).
Effect of MYC induction on age-associated gene expression program
To test the specificity of the “rejuvenation” effect of NF-κB blockade, we examined whether enforced cell cycle entry by activation of the K14:MYC-ER transgene (Frye et al. 2003) can induce the same gene expression program. We averaged the expression of genes that are induced by age and reversed by NF-κB blockade in 18- to 23-mo-old K14:NFKB1ΔSP-ER mice. We used Unigene identifier to match genes represented in different microarray platforms. Of the 225 “rejuvenated” genes, 70 were in common between Stanford cDNA arrays and Affymetrix Genechips and were analyzed. The average expression levels of these genes were determined in K14:MYC-ER mice in the presence of 4-OHT for 1 or 4 d relative to samples in the absence of 4-OHT treatment. Expression levels of the KRT2-6a (Keratin 6) and KRT1-17 (Keratin 17) genes were extracted from the microarray data and compared by Student’s t-test.
Effect of hyperproliferation on age-associated gene expression program
To determine the effect of hyperproliferation on the aging signature in the skin of old mice, we compared the average expression of all 225 “rejuvenated” genes following acetone or TPA treatment of old wild-type skin.
Comparison of NFKB1ΔSP-ER and IκBα-SR repressors on TNF-α-induced genes
SAM between TNF-α-induced (10 ng/mL, 1.5 h) primary human keratinocytes expressing GFP or NFKB1ΔSP-ER + EtOH and cells expressing IκBα-SR identified genes that were induced by TNF-α but repressed by expression of IκBα-SR (FDR < 0.03). The activity of these genes following introduction of NFKB1ΔSP-ER + 4-OHT (500 nM, 21 h) was analyzed. Motif module map was used to identify the average expression levels of specific motif modules in these same samples.
Histological and immunocytochemical analyses
Immediately after harvesting, skin samples were fixed in formalin. Sections were embedded in paraffin, sectioned, and stained for hematoxylin and eosin by Histo-Tec Laboratories. The thickness of the epidermis (number of cell layers) was quantified every 250 μm of epidermis for a minimum of 8.5 mm for each skin sample and then averaged. Immunohistochemical staining for p16INK4A, Ki-67, and cleaved Caspase-3 protein in paraffin-embedded sections (5 μm) was performed as described (J.Y. Zhang et al. 2004) using BioGenex IHC detection systems. Anti-p16INK4A (F-12; Santa Cruz Biotechnology), anti-Ki-67 (TEC-3; DakoCytomation), and anti-cleaved Caspase-3 (Asp175; Cell Signaling Technology) were from the indicated sources. Senescence-associated β-galactosidase (SA-β-gal) activity was detected with a SA-β-gal staining kit (Cell Signaling Technology) in OCT-embedded frozen sections (6 μm). Blue color was developed by incubation overnight at 37°C. Relative protein expression and SA-β-gal activity was quantified on a 0–3 scale: 0 indicates no staining; 1 signifies weak staining; 2 indicates intermediate staining; and 3 signifies strong staining. A score was obtained every 250 μm (p16INK4A) or 175 μm (SA-β-gal) of epidermis (minimum of 8.5 mm or 8.75 mm, respectively) for each sample and then averaged. The number of Ki-67-positive cells was counted every 500 μm of epidermis for a minimum of 8 mm for each sample and then averaged. Immunofluorescence of K14, K10, and loricrin protein in OCT-embedded frozen sections (6 μm) and p50 in primary human keratinocytes was performed as described (Zhang et al. 2005). Anti-K14 (AF 64; Covance), anti-K10 (Covance), anti-loricrin (AF 62; Covance), anti-p50 (C-19, Santa Cruz Biotechnology), and Cy3-conjugated IgG (Jackson ImmunoResearch Laboratories, Inc.) were from the indicated sources.
URLs
Microarray data are available at Stanford Microarray Database (http://smd.stanford.edu) and GEO (http://www.ncbi.nlm.nih.gov/geo; GSE3845).
Acknowledgments
We thank P.A. Khavari, F.A. Scholl, A. Brunet, and V. Beaudry for reagents and support; P.O. Brown, P.A. Khavari, S.K. Kim, F. Watt, and C.S. April for microarray data; and D. Baltimore, P.A. Khavari, A. Hoffmann, and S.K. Kim for critical review of the manuscript. Work was supported by grants from the American Cancer Society grant RSG 07-084-01 (to H.Y.C.); National Institutes of Health grant K08-AR050007 (to H.Y.C.); Israel Science Foundation (to E.S.); the National Cancer Institute, DHHS (to A.S.A.); and the California Breast Cancer Research Program (to H.Y.C. and A.S.A.). E.S. is the incumbent of the Soretta and Henry Shapiro career development chair; H.Y.C. is the Kenneth G. and Elaine A. Langone Scholar supported by the Damon Runyon Cancer Research Foundation.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1588507
References
- April C.S., Barsh G.S., Barsh G.S. Skin layer-specific transcriptional profiles in normal and recessive yellow (Mc1re/Mc1re) mice. Pigment Cell Res. 2006;19:194–205. doi: 10.1111/j.1600-0749.2006.00305.x. [DOI] [PubMed] [Google Scholar]
- Arkan M.C., Hevener A.L., Greten F.R., Maeda S., Li Z.W., Long J.M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Hevener A.L., Greten F.R., Maeda S., Li Z.W., Long J.M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Greten F.R., Maeda S., Li Z.W., Long J.M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Maeda S., Li Z.W., Long J.M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Li Z.W., Long J.M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Long J.M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Wynshaw-Boris A., Poli G., Olefsky J., Karin M., Poli G., Olefsky J., Karin M., Olefsky J., Karin M., Karin M. IKK-β links inflammation to obesity-induced insulin resistance. Nat. Med. 2005;11:191–198. doi: 10.1038/nm1185. [DOI] [PubMed] [Google Scholar]
- Baur J.A., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Allard J.S., Lopez-Lluch G., Lewis K., Lopez-Lluch G., Lewis K., Lewis K., et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard D., Gosselin K., Monte D., Vercamer C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C., Gosselin K., Monte D., Vercamer C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C., Monte D., Vercamer C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C., Vercamer C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C., Bouali F., Pourtier A., Vandenbunder B., Abbadie C., Pourtier A., Vandenbunder B., Abbadie C., Vandenbunder B., Abbadie C., Abbadie C. Involvement of Rel/nuclear factor-κB transcription factors in keratinocyte senescence. Cancer Res. 2004;64:472–481. doi: 10.1158/0008-5472.can-03-0005. [DOI] [PubMed] [Google Scholar]
- Borneman A.R., Leigh-Bell J.A., Yu H., Bertone P., Gerstein M., Snyder M., Leigh-Bell J.A., Yu H., Bertone P., Gerstein M., Snyder M., Yu H., Bertone P., Gerstein M., Snyder M., Bertone P., Gerstein M., Snyder M., Gerstein M., Snyder M., Snyder M. Target hub proteins serve as master regulators of development in yeast. Genes & Dev. 2006;20:435–448. doi: 10.1101/gad.1389306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai D., Frantz J.D., Tawa N.E., Melendez P.A., Oh B.C., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Frantz J.D., Tawa N.E., Melendez P.A., Oh B.C., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Tawa N.E., Melendez P.A., Oh B.C., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Melendez P.A., Oh B.C., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Oh B.C., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Lidov H.G., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Hasselgren P.O., Frontera W.R., Lee J., Glass D.J., Frontera W.R., Lee J., Glass D.J., Lee J., Glass D.J., Glass D.J., et al. IKKβ/NF-κB activation causes severe muscle wasting in mice. Cell. 2004;119:285–298. doi: 10.1016/j.cell.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Chang H.Y., Chi J.T., Dudoit S., Bondre C., de van Rijn M., Botstein D., Brown P.O., Chi J.T., Dudoit S., Bondre C., de van Rijn M., Botstein D., Brown P.O., Dudoit S., Bondre C., de van Rijn M., Botstein D., Brown P.O., Bondre C., de van Rijn M., Botstein D., Brown P.O., de van Rijn M., Botstein D., Brown P.O., Botstein D., Brown P.O., Brown P.O. Diversity, topographic differentiation, and positional memory in human fibroblasts. Proc. Natl. Acad. Sci. 2002;99:12877–12882. doi: 10.1073/pnas.162488599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Zhou Y., Mueller-Steiner S., Chen L.F., Kwon H., Yi S., Mucke L., Gan L., Zhou Y., Mueller-Steiner S., Chen L.F., Kwon H., Yi S., Mucke L., Gan L., Mueller-Steiner S., Chen L.F., Kwon H., Yi S., Mucke L., Gan L., Chen L.F., Kwon H., Yi S., Mucke L., Gan L., Kwon H., Yi S., Mucke L., Gan L., Yi S., Mucke L., Gan L., Mucke L., Gan L., Gan L. SIRT1 Protects against microglia-dependent amyloid-β toxicity through inhibiting NF-κB signaling. J. Biol. Chem. 2005;280:40364–40374. doi: 10.1074/jbc.M509329200. [DOI] [PubMed] [Google Scholar]
- Chung H.Y., Kim H.J., Kim J.W., Yu B.P., Kim H.J., Kim J.W., Yu B.P., Kim J.W., Yu B.P., Yu B.P. The inflammation hypothesis of aging: Molecular modulation by calorie restriction. Ann. N. Y. Acad. Sci. 2001;928:327–335. [PubMed] [Google Scholar]
- Conboy I.M., Conboy M.J., Wagers A.J., Girma E.R., Weissman I.L., Rando T.A., Conboy M.J., Wagers A.J., Girma E.R., Weissman I.L., Rando T.A., Wagers A.J., Girma E.R., Weissman I.L., Rando T.A., Girma E.R., Weissman I.L., Rando T.A., Weissman I.L., Rando T.A., Rando T.A. Rejuvenation of aged progenitor cells by exposure to a young systemic environment. Nature. 2005;433:760–764. doi: 10.1038/nature03260. [DOI] [PubMed] [Google Scholar]
- Dhahbi J.M., Kim H.J., Mote P.L., Beaver R.J., Spindler S.R., Kim H.J., Mote P.L., Beaver R.J., Spindler S.R., Mote P.L., Beaver R.J., Spindler S.R., Beaver R.J., Spindler S.R., Spindler S.R. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc. Natl. Acad. Sci. 2004;101:5524–5529. doi: 10.1073/pnas.0305300101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., Linskens M., Rubelj I., Pereira-Smith O., Rubelj I., Pereira-Smith O., Pereira-Smith O., et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.B., Khaitovich P., Plotkin J.B., Paabo S., Eisen M.B., Khaitovich P., Plotkin J.B., Paabo S., Eisen M.B., Plotkin J.B., Paabo S., Eisen M.B., Paabo S., Eisen M.B., Eisen M.B. Aging and gene expression in the primate brain. PLoS Biol. 2005;3:e274. doi: 10.1371/journal.pbio.0030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye M., Gardner C., Li E.R., Arnold I., Watt F.M., Gardner C., Li E.R., Arnold I., Watt F.M., Li E.R., Arnold I., Watt F.M., Arnold I., Watt F.M., Watt F.M. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- Funk W.D., Wang C.K., Shelton D.N., Harley C.B., Pagon G.D., Hoeffler W.K., Wang C.K., Shelton D.N., Harley C.B., Pagon G.D., Hoeffler W.K., Shelton D.N., Harley C.B., Pagon G.D., Hoeffler W.K., Harley C.B., Pagon G.D., Hoeffler W.K., Pagon G.D., Hoeffler W.K., Hoeffler W.K. Telomerase expression restores dermal integrity to in vitro-aged fibroblasts in a reconstituted skin model. Exp. Cell Res. 2000;258:270–278. doi: 10.1006/excr.2000.4945. [DOI] [PubMed] [Google Scholar]
- Hardy K., Mansfield L., Mackay A., Benvenuti S., Ismail S., Arora P., O’Hare M.J., Jat P.S., Mansfield L., Mackay A., Benvenuti S., Ismail S., Arora P., O’Hare M.J., Jat P.S., Mackay A., Benvenuti S., Ismail S., Arora P., O’Hare M.J., Jat P.S., Benvenuti S., Ismail S., Arora P., O’Hare M.J., Jat P.S., Ismail S., Arora P., O’Hare M.J., Jat P.S., Arora P., O’Hare M.J., Jat P.S., O’Hare M.J., Jat P.S., Jat P.S. Transcriptional networks and cellular senescence in human mammary fibroblasts. Mol. Biol. Cell. 2005;16:943–953. doi: 10.1091/mbc.E04-05-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S., Ghosh S. Signaling to NF-κB. Genes & Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hinata K., Gervin A.M., Zhang J.Y., Khavari P.A., Gervin A.M., Zhang J.Y., Khavari P.A., Zhang J.Y., Khavari P.A., Khavari P.A. Divergent gene regulation and growth effects by NF-κB in epithelial and mesenchymal cells of human skin. Oncogene. 2003;22:1955–1964. doi: 10.1038/sj.onc.1206198. [DOI] [PubMed] [Google Scholar]
- Hu M.C., Lee D.F., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Lee D.F., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., Zou Y., Bao S., Hanada N., Saso H., Bao S., Hanada N., Saso H., Hanada N., Saso H., Saso H., et al. IκB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J., Torrice C., Ramsey M.R., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E., Torrice C., Ramsey M.R., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E., Ramsey M.R., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E., Al-Regaiey K., Su L., Sharpless N.E., Su L., Sharpless N.E., Sharpless N.E. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C.A., Yao F., Wong J.J., George J., Xu H., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., Yao F., Wong J.J., George J., Xu H., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., Wong J.J., George J., Xu H., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., George J., Xu H., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., Xu H., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., Chiu K.P., Sung W.K., Lipovich L., Vega V.B., Chen J., Sung W.K., Lipovich L., Vega V.B., Chen J., Lipovich L., Vega V.B., Chen J., Vega V.B., Chen J., Chen J., et al. Genome-wide mapping of RELA(p65) binding identifies E2F1 as a transcriptional activator recruited by NF-κB upon TLR4 activation. Mol. Cell. 2007;27:622–635. doi: 10.1016/j.molcel.2007.06.038. [DOI] [PubMed] [Google Scholar]
- Lin L., Hron J.D., Peng S.L., Hron J.D., Peng S.L., Peng S.L. Regulation of NF-κB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Logeat F., Israel N., Ten R., Blank V., Le Bail O., Kourilsky P., Israel A., Israel N., Ten R., Blank V., Le Bail O., Kourilsky P., Israel A., Ten R., Blank V., Le Bail O., Kourilsky P., Israel A., Blank V., Le Bail O., Kourilsky P., Israel A., Le Bail O., Kourilsky P., Israel A., Kourilsky P., Israel A., Israel A. Inhibition of transcription factors belonging to the rel/NF-κB family by a transdominant negative mutant. EMBO J. 1991;10:1827–1832. doi: 10.1002/j.1460-2075.1991.tb07708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T., Pan Y., Kao S.Y., Li C., Kohane I., Chan J., Yankner B.A., Pan Y., Kao S.Y., Li C., Kohane I., Chan J., Yankner B.A., Kao S.Y., Li C., Kohane I., Chan J., Yankner B.A., Li C., Kohane I., Chan J., Yankner B.A., Kohane I., Chan J., Yankner B.A., Chan J., Yankner B.A., Yankner B.A. Gene regulation and DNA damage in the ageing human brain. Nature. 2004;429:883–891. doi: 10.1038/nature02661. [DOI] [PubMed] [Google Scholar]
- Mair W., Goymer P., Pletcher S.D., Partridge L., Goymer P., Pletcher S.D., Partridge L., Pletcher S.D., Partridge L., Partridge L. Demography of dietary restriction and death in Drosophila. Science. 2003;301:1731–1733. doi: 10.1126/science.1086016. [DOI] [PubMed] [Google Scholar]
- Matys V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Fricke E., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Geffers R., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Gossling E., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Haubrock M., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hehl R., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Hornischer K., Karas D., Kel A.E., Kel-Margoulis O.V., Karas D., Kel A.E., Kel-Margoulis O.V., Kel A.E., Kel-Margoulis O.V., Kel-Margoulis O.V., et al. TRANSFAC: Transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarroll S.A., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Murphy C.T., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Zou S., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Pletcher S.D., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Chin C.S., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Jan Y.N., Kenyon C., Bargmann C.I., Li H., Kenyon C., Bargmann C.I., Li H., Bargmann C.I., Li H., Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nat. Genet. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- Melov S., Tarnopolsky M.A., Beckman K., Felkey K., Hubbard A., Tarnopolsky M.A., Beckman K., Felkey K., Hubbard A., Beckman K., Felkey K., Hubbard A., Felkey K., Hubbard A., Hubbard A. Resistance exercise reverses aging in human skeletal muscle. PLoS ONE. 2007;2:e465. doi: 10.1371/journal.pone.0000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki R., Kadota K., Bono H., Mizuno Y., Tomaru Y., Carninci P., Itoh M., Shibata K., Kawai J., Konno H., Kadota K., Bono H., Mizuno Y., Tomaru Y., Carninci P., Itoh M., Shibata K., Kawai J., Konno H., Bono H., Mizuno Y., Tomaru Y., Carninci P., Itoh M., Shibata K., Kawai J., Konno H., Mizuno Y., Tomaru Y., Carninci P., Itoh M., Shibata K., Kawai J., Konno H., Tomaru Y., Carninci P., Itoh M., Shibata K., Kawai J., Konno H., Carninci P., Itoh M., Shibata K., Kawai J., Konno H., Itoh M., Shibata K., Kawai J., Konno H., Shibata K., Kawai J., Konno H., Kawai J., Konno H., Konno H., et al. Delineating developmental and metabolic pathways in vivo by expression profiling using the RIKEN set of 18,816 full-length enriched mouse cDNA arrays. Proc. Natl. Acad. Sci. 2001;98:2199–2204. doi: 10.1073/pnas.041605498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C.T., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., McCarroll S.A., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., Bargmann C.I., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., Fraser A., Kamath R.S., Ahringer J., Li H., Kenyon C., Kamath R.S., Ahringer J., Li H., Kenyon C., Ahringer J., Li H., Kenyon C., Li H., Kenyon C., Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Murray J.I., Whitfield M.L., Trinklein N.D., Myers R.M., Brown P.O., Botstein D., Whitfield M.L., Trinklein N.D., Myers R.M., Brown P.O., Botstein D., Trinklein N.D., Myers R.M., Brown P.O., Botstein D., Myers R.M., Brown P.O., Botstein D., Brown P.O., Botstein D., Botstein D. Diverse and specific gene expression responses to stresses in cultured human cells. Mol. Biol. Cell. 2004;15:2361–2374. doi: 10.1091/mbc.E03-11-0799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed T., Dick D.M., Uniacke S.K., Foroud T., Nichols W.C., Dick D.M., Uniacke S.K., Foroud T., Nichols W.C., Uniacke S.K., Foroud T., Nichols W.C., Foroud T., Nichols W.C., Nichols W.C. Genome-wide scan for a healthy aging phenotype provides support for a locus near D4S1564 promoting healthy aging. J. Gerontol. A Biol. Sci. Med. Sci. 2004;59:227–232. doi: 10.1093/gerona/59.3.b227. [DOI] [PubMed] [Google Scholar]
- Segal E., Friedman N., Kaminski N., Regev A., Koller D., Friedman N., Kaminski N., Regev A., Koller D., Kaminski N., Regev A., Koller D., Regev A., Koller D., Koller D. From signatures to models: Understanding cancer using microarrays. Nat. Genet. 2005;37 (Suppl.):S38–S45. doi: 10.1038/ng1561. [DOI] [PubMed] [Google Scholar]
- Sinha S., van Nimwegen E., Siggia E.D., van Nimwegen E., Siggia E.D., Siggia E.D. A probabilistic method to detect regulatory modules. Bioinformatics. 2003;19 (Suppl. 1):i292–i301. doi: 10.1093/bioinformatics/btg1040. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Pomeroy S.L., Golub T.R., Lander E.S., Golub T.R., Lander E.S., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G., Tibshirani R., Chu G., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung F., Hoberg J.E., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A., Mayo M.W., Hoberg J.E., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A., Mayo M.W., Ramsey C.S., Keller M.D., Jones D.R., Frye R.A., Mayo M.W., Keller M.D., Jones D.R., Frye R.A., Mayo M.W., Jones D.R., Frye R.A., Mayo M.W., Frye R.A., Mayo M.W., Mayo M.W. Modulation of NF-κB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H.G., Hyde K., Page G.P., Brand J.P., Zhou J., Yu S., Allison D.B., Hsu H.C., Mountz J.D., Hyde K., Page G.P., Brand J.P., Zhou J., Yu S., Allison D.B., Hsu H.C., Mountz J.D., Page G.P., Brand J.P., Zhou J., Yu S., Allison D.B., Hsu H.C., Mountz J.D., Brand J.P., Zhou J., Yu S., Allison D.B., Hsu H.C., Mountz J.D., Zhou J., Yu S., Allison D.B., Hsu H.C., Mountz J.D., Yu S., Allison D.B., Hsu H.C., Mountz J.D., Allison D.B., Hsu H.C., Mountz J.D., Hsu H.C., Mountz J.D., Mountz J.D. Novel tumor necrosis factor α-regulated genes in rheumatoid arthritis. Arthritis Rheum. 2004;50:420–431. doi: 10.1002/art.20037. [DOI] [PubMed] [Google Scholar]
- Zhang J.Y., Green C.L., Tao S., Khavari P.A., Green C.L., Tao S., Khavari P.A., Tao S., Khavari P.A., Khavari P.A. NF-κB RelA opposes epidermal proliferation driven by TNFR1 and JNK. Genes & Dev. 2004;18:17–22. doi: 10.1101/gad.1160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.Y., Tao S., Kimmel R., Khavari P.A., Tao S., Kimmel R., Khavari P.A., Kimmel R., Khavari P.A., Khavari P.A. CDK4 regulation by TNFR1 and JNK is required for NF-κB-mediated epidermal growth control. J. Cell Biol. 2005;168:561–566. doi: 10.1083/jcb.200411060. [DOI] [PMC free article] [PubMed] [Google Scholar]