Abstract
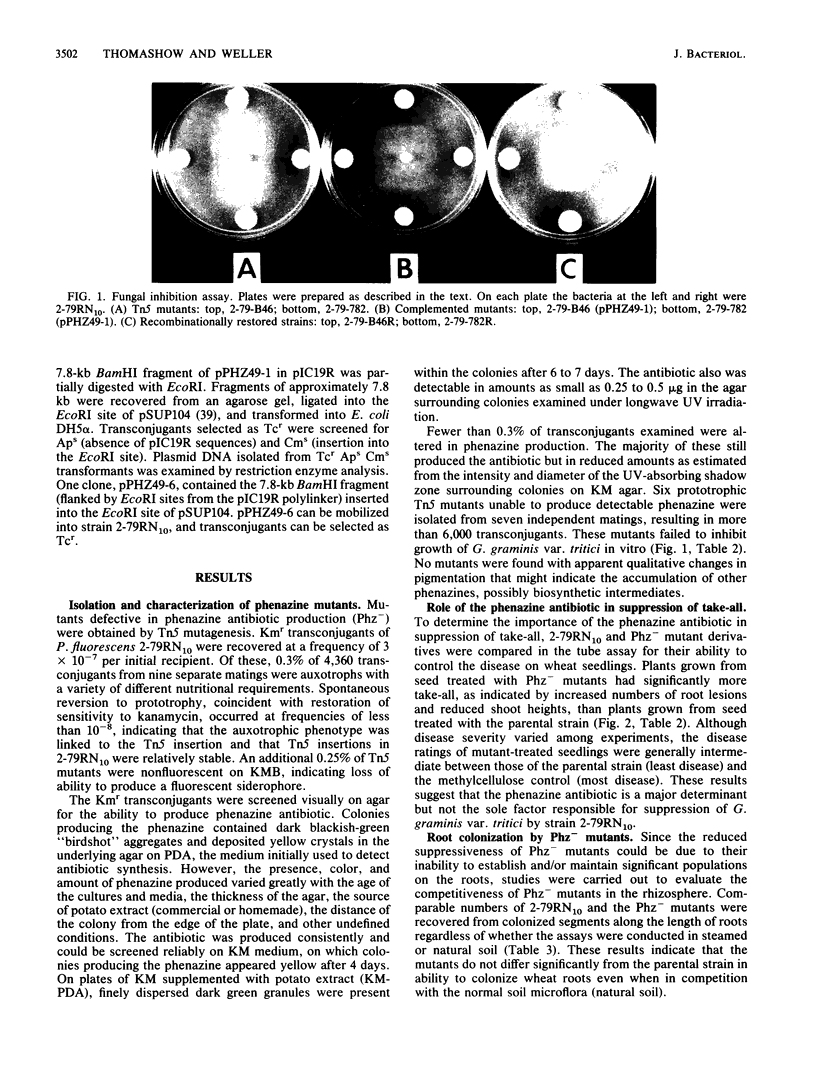
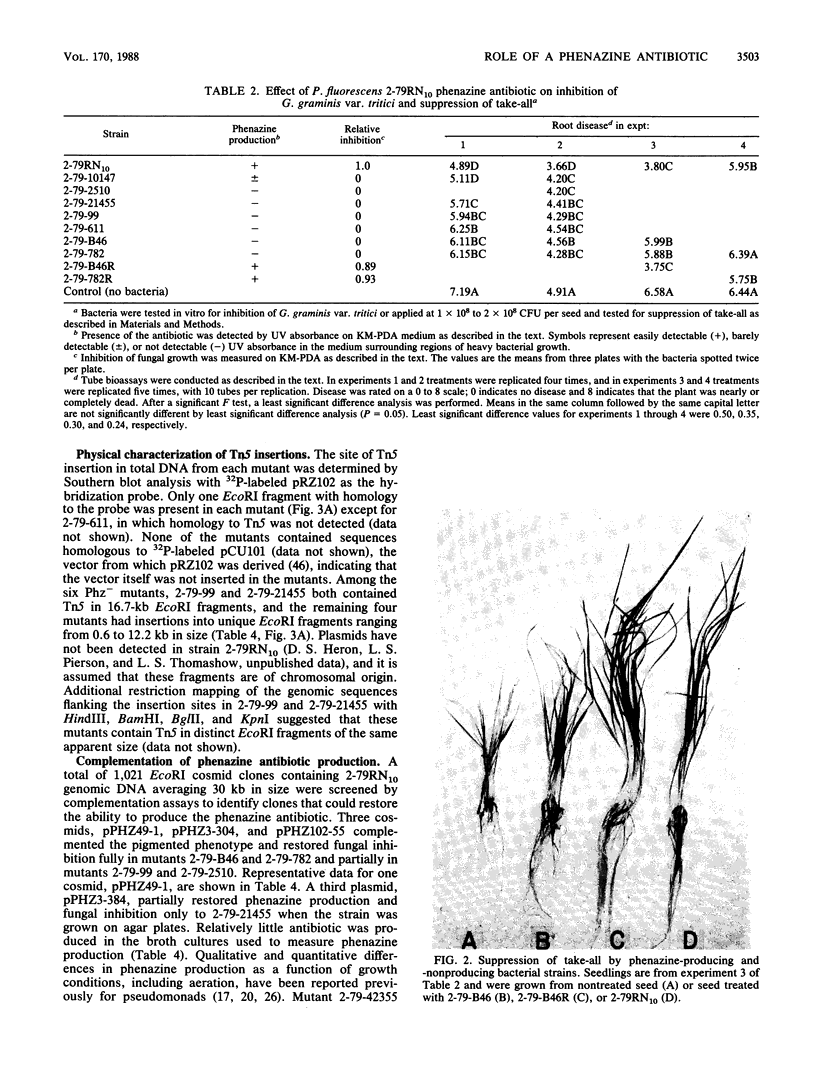
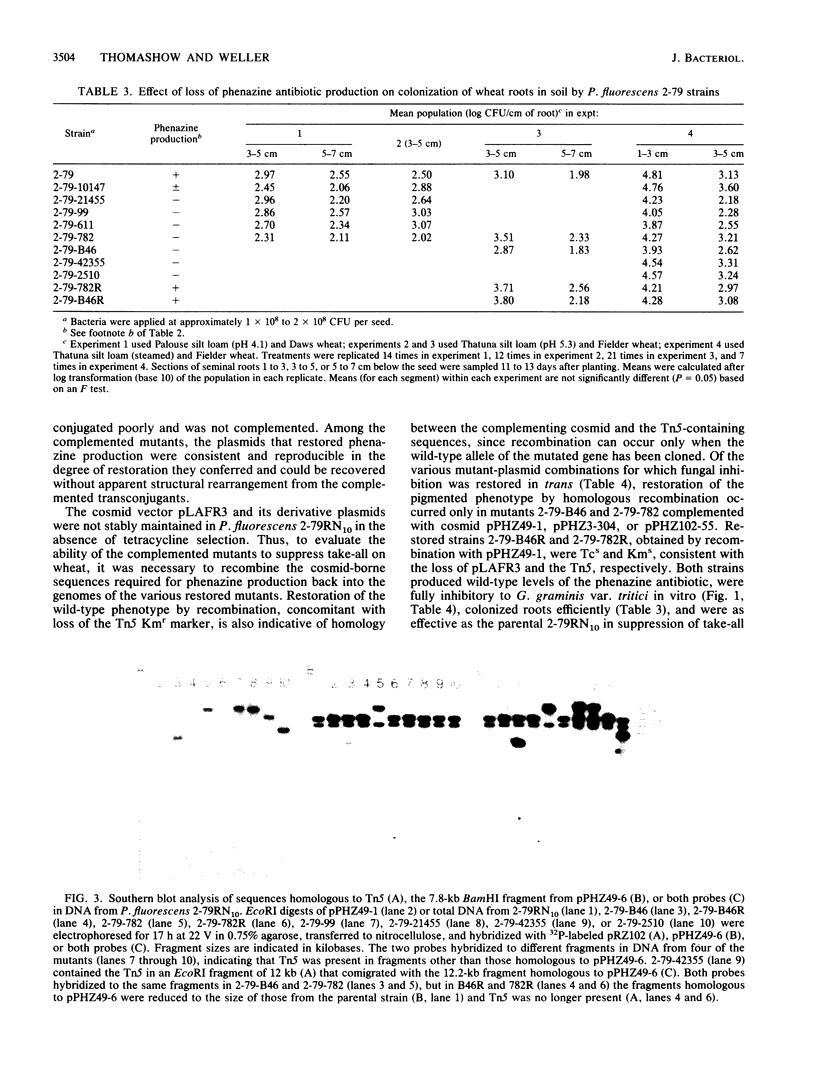
Pseudomonas fluorescens 2-79 (NRRL B-15132) and its rifampin-resistant derivative 2-79RN10 are suppressive to take-all, a major root disease of wheat caused by Gaeumannomyces graminis var. tritici. Strain 2-79 produces the antibiotic phenazine-1-carboxylate, which is active in vitro against G. graminis var. tritici and other fungal root pathogens. Mutants defective in phenazine synthesis (Phz-) were generated by Tn5 insertion and then compared with the parental strain to determine the importance of the antibiotic in take-all suppression on wheat roots. Six independent, prototrophic Phz- mutants were noninhibitory to G. graminis var. tritici in vitro and provided significantly less control of take-all than strain 2-79 on wheat seedlings. Antibiotic synthesis, fungal inhibition in vitro, and suppression of take-all on wheat were coordinately restored in two mutants complemented with cloned DNA from a 2-79 genomic library. These mutants contained Tn5 insertions in adjacent EcoRI fragments in the 2-79 genome, and the restriction maps of the region flanking the insertions and the complementary DNA were colinear. These results indicate that sequences required for phenazine production were present in the cloned DNA and support the importance of the phenazine antibiotic in disease suppression in the rhizosphere.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brisbane P. G., Janik L. J., Tate M. E., Warren R. F. Revised structure for the phenazine antibiotic from Pseudomonas fluorescens 2-79 (NRRL B-15132). Antimicrob Agents Chemother. 1987 Dec;31(12):1967–1971. doi: 10.1128/aac.31.12.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byng G. S., Berry A., Jensen R. A. A pair of regulatory isozymes for 3-deoxy-D-arabino-heptulosonate 7-phosphate synthase is conserved within group I pseudomonads. J Bacteriol. 1983 Oct;156(1):429–433. doi: 10.1128/jb.156.1.429-433.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byng G. S., Jensen R. A. Impact of isozymes upon partitioning of carbon flow and regulation of aromatic biosynthesis in prokaryotes. Isozymes Curr Top Biol Med Res. 1983;8:115–140. [PubMed] [Google Scholar]
- Byng G. S., Johnson J. L., Whitaker R. J., Gherna R. L., Jensen R. A. The evolutionary pattern of aromatic amino acid biosynthesis and the emerging phylogeny of pseudomonad bacteria. J Mol Evol. 1983;19(3-4):272–282. doi: 10.1007/BF02099974. [DOI] [PubMed] [Google Scholar]
- Calhoun D. H., Carson M., Jensen R. A. The branch point metabolite for pyocyanine biosynthesis in Pseudomonas aeruginosa. J Gen Microbiol. 1972 Oct;72(3):581–583. doi: 10.1099/00221287-72-3-581. [DOI] [PubMed] [Google Scholar]
- Currier T. C., Nester E. W. Isolation of covalently closed circular DNA of high molecular weight from bacteria. Anal Biochem. 1976 Dec;76(2):431–441. doi: 10.1016/0003-2697(76)90338-9. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurusiddaiah S., Weller D. M., Sarkar A., Cook R. J. Characterization of an antibiotic produced by a strain of Pseudomonas fluorescens inhibitory to Gaeumannomyces graminis var. tritici and Pythium spp. Antimicrob Agents Chemother. 1986 Mar;29(3):488–495. doi: 10.1128/aac.29.3.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutterson N. I., Layton T. J., Ziegle J. S., Warren G. J. Molecular cloning of genetic determinants for inhibition of fungal growth by a fluorescent pseudomonad. J Bacteriol. 1986 Mar;165(3):696–703. doi: 10.1128/jb.165.3.696-703.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliman F. G. Pigments of pseudomonas species. I. Structure and synthesis of aeruginosin A. J Chem Soc Perkin 1. 1969;18:2514–2516. doi: 10.1039/j39690002514. [DOI] [PubMed] [Google Scholar]
- James D. W., Jr, Gutterson N. I. Multiple antibiotics produced by Pseudomonas fluorescens HV37a and their differential regulation by glucose. Appl Environ Microbiol. 1986 Nov;52(5):1183–1189. doi: 10.1128/aem.52.5.1183-1189.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Kanner D., Gerber N. N., Bartha R. Pattern of phenazine pigment production by a strain of Pseudomonas aeruginosa. J Bacteriol. 1978 May;134(2):690–692. doi: 10.1128/jb.134.2.690-692.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitch M. E. Regulation of aromatic amino acid biosynthesis in phenazine-producing strains of Pseudomonas. J Bacteriol. 1970 Jul;103(1):16–19. doi: 10.1128/jb.103.1.16-19.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley R. P., Halliwell J. E., Campbell J. J., Ingledew W. M. The branchpoint of pyocyanine biosynthesis. Can J Microbiol. 1972 Sep;18(9):1357–1363. doi: 10.1139/m72-210. [DOI] [PubMed] [Google Scholar]
- Marsh J. L., Erfle M., Wykes E. J. The pIC plasmid and phage vectors with versatile cloning sites for recombinant selection by insertional inactivation. Gene. 1984 Dec;32(3):481–485. doi: 10.1016/0378-1119(84)90022-2. [DOI] [PubMed] [Google Scholar]
- Marugg J. D., van Spanje M., Hoekstra W. P., Schippers B., Weisbeek P. J. Isolation and analysis of genes involved in siderophore biosynthesis in plant-growth-stimulating Pseudomonas putida WCS358. J Bacteriol. 1985 Nov;164(2):563–570. doi: 10.1128/jb.164.2.563-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peet R. C., Lindgren P. B., Willis D. K., Panopoulos N. J. Identification and cloning of genes involved in phaseolotoxin production by Pseudomonas syringae pv. "phaseolicola". J Bacteriol. 1986 Jun;166(3):1096–1105. doi: 10.1128/jb.166.3.1096-1105.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefer U. B., Simon R., Pühler A. Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol. 1985 Jul;163(1):324–330. doi: 10.1128/jb.163.1.324-330.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroth M. N., Hancock J. G. Disease-suppressive soil and root-colonizing bacteria. Science. 1982 Jun 25;216(4553):1376–1381. doi: 10.1126/science.216.4553.1376. [DOI] [PubMed] [Google Scholar]
- Schroth M. N., Hancock J. G. Selected topics in biological control. Annu Rev Microbiol. 1981;35:453–476. doi: 10.1146/annurev.mi.35.100181.002321. [DOI] [PubMed] [Google Scholar]
- Selvaraj G., Iyer V. N. Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol. 1983 Dec;156(3):1292–1300. doi: 10.1128/jb.156.3.1292-1300.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker R. J., Byng G. S., Gherna R. L., Jensen R. A. Comparative allostery of 3-deoxy-D-arabino-heptulosonate 7-phosphate synthetase as an indicator of taxonomic relatedness in pseudomonad genera. J Bacteriol. 1981 Feb;145(2):752–759. doi: 10.1128/jb.145.2.752-759.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]





