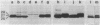Abstract
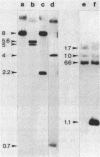
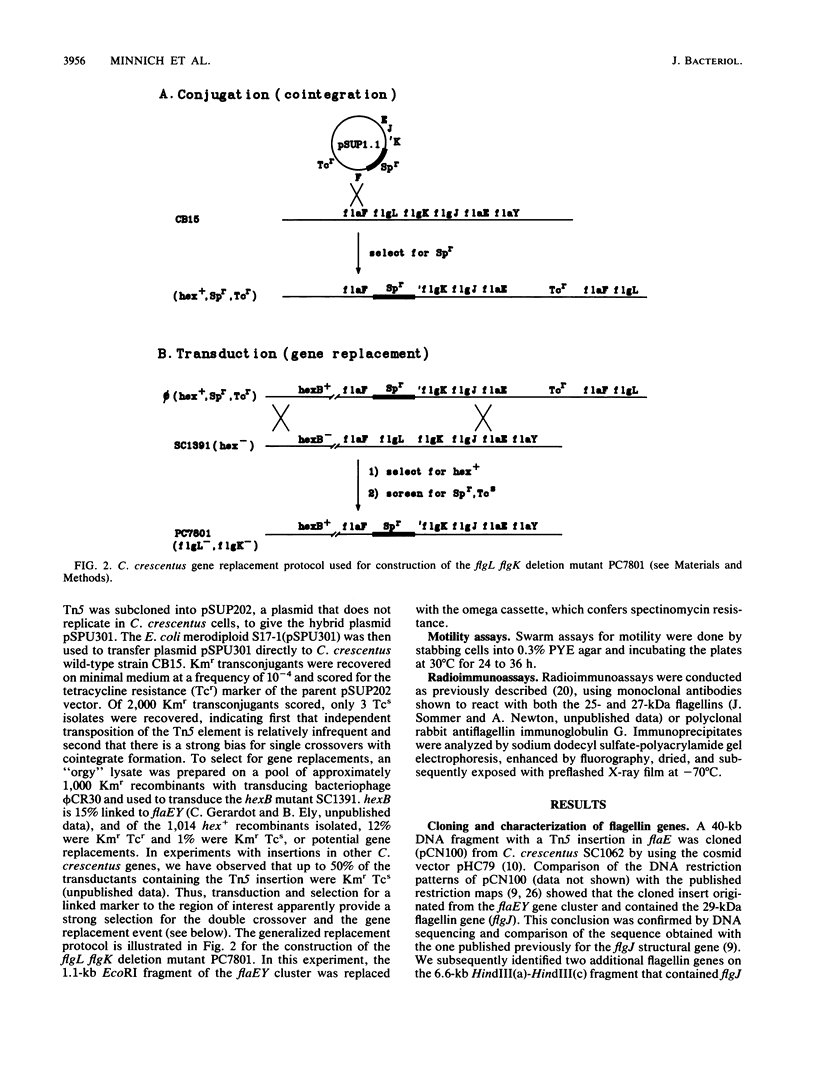
Caulobacter crescentus incorporates two distinct, but related proteins into the polar flagellar filament: a 27-kilodalton (kDa) flagellin is assembled proximal to the hook and a 25-kDa flagellin forms the distal end of the filament. These two proteins and a third, related flagellin protein of 29 kDa are encoded by three tandem genes (alpha-flagellin cluster) in the flaEY gene cluster (S.A. Minnich and A. Newton, Proc. Natl. Acad. Sci. USA 84: 1142-1146, 1987). Since point mutations in flagellin genes had not been isolated their requirement for flagellum function and fla gene expression was not known. To address these questions, we developed a gene replacement protocol that uses cloned flagellin genes mutagenized by either Tn5 transposons in vivo or the replacement of specific DNA fragments in vitro by the antibiotic resistance omega cassette. Analysis of gene replacement mutants constructed by this procedure led to several conclusions. (i) Mutations in any of the three flagellin genes do not cause complete loss of motility. (ii) Tn5 insertions in the 27-kDa flagellin gene and a deletion mutant of this gene do not synthesize the 27-kDa flagellin, but they do synthesize wild-type levels of the 25-kDa flagellin, which implies that the 27-kDa flagellin is not required for expression and assembly of the 25-kDa flagellin; these mutants show slightly impaired motility on swarm plates. (iii) Mutant PC7810, which is deleted for the three flagellin genes in the flaEY cluster, does not synthesize the 27- or 29-kDa flagellin, and it is significantly more impaired for motility on swarm plates than mutants with defects in only the 27-kDa flagellin gene. The synthesis of essentially normal levels of 25-kDa flagellin by strain PC7810 confirms that additional copies of the 25-kDa flagellin map outside the flaEY cluster (beta-flagellin cluster) and that these flagellin genes are active. Thus, while the 29- and 27-kDa flagellins are not absolutely essential for motility in C. crescentus, their assembly into the flagellar structure is necessary for normal flagellar function.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrett J. T., Rhodes C. S., Ferber D. M., Jenkins B., Kuhl S. A., Ely B. Construction of a genetic map for Caulobacter crescentus. J Bacteriol. 1982 Mar;149(3):889–896. doi: 10.1128/jb.149.3.889-896.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan R., Purucker M., Gomes S. L., Alexander W., Shapiro L. Analysis of the pleiotropic regulation of flagellar and chemotaxis gene expression in Caulobacter crescentus by using plasmid complementation. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1341–1345. doi: 10.1073/pnas.81.5.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. S., Mullin D., Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci U S A. 1986 May;83(9):2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H., Gerardot C. J. Genetic mapping of genes required for motility in Caulobacter crescentus. Genetics. 1984 Nov;108(3):523–532. doi: 10.1093/genetics/108.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Johnson R. C. Generalized Transduction in CAULOBACTER CRESCENTUS. Genetics. 1977 Nov;87(3):391–399. doi: 10.1093/genetics/87.3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P. R., Agabian N. The nucleotide sequence of the Mr = 28,500 flagellin gene of Caulobacter crescentus. J Biol Chem. 1983 Jun 25;258(12):7395–7401. [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Komeda Y. Fusions of flagellar operons to lactose genes on a mu lac bacteriophage. J Bacteriol. 1982 Apr;150(1):16–26. doi: 10.1128/jb.150.1.16-26.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyasu S., Asada M., Fukuda A., Okada Y. Sequential polymerization of flagellin A and flagellin B into Caulobacter flagella. J Mol Biol. 1981 Dec 5;153(2):471–475. doi: 10.1016/0022-2836(81)90292-8. [DOI] [PubMed] [Google Scholar]
- Lagenaur C., Agabian N. Caulobacter flagellar organelle: synthesis, compartmentation, and assembly. J Bacteriol. 1978 Sep;135(3):1062–1069. doi: 10.1128/jb.135.3.1062-1069.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milhausen M., Gill P. R., Parker G., Agabian N. Cloning of developmentally regulated flagellin genes from Caulobacter crescentus via immunoprecipitation of polyribosomes. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6847–6851. doi: 10.1073/pnas.79.22.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich S. A., Newton A. Promoter mapping and cell cycle regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton A., Allebach E. Gene transfer in Caulobacter crescentus: polarized inheritance of genetic markers. Genetics. 1975 May;80(1):1–11. doi: 10.1093/genetics/80.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Newton A. Isolation and expression of cloned hook protein gene from Caulobacter crescentus. Proc Natl Acad Sci U S A. 1982 Aug;79(16):4863–4867. doi: 10.1073/pnas.79.16.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Swanson E., Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985 Nov 5;186(1):107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- Ohta N., Swanson E., Ely B., Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984 Jun;158(3):897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Sheffery M., Newton A. Regulation of flagellin synthesis in the cell cycle of caulobacter: dependence on DNA replication. Cell. 1977 Oct;12(2):393–400. doi: 10.1016/0092-8674(77)90115-5. [DOI] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentki P., Krisch H. M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984 Sep;29(3):303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- Purucker M., Bryan R., Amemiya K., Ely B., Shapiro L. Isolation of a Caulobacter gene cluster specifying flagellum production by using nonmotile Tn5 insertion mutants. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6797–6801. doi: 10.1073/pnas.79.22.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Shapiro L. Generation of polarity during Caulobacter cell differentiation. Annu Rev Cell Biol. 1985;1:173–207. doi: 10.1146/annurev.cb.01.110185.001133. [DOI] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Reconstitution and purification of flagellar filaments from Caulobacter crescentus. J Bacteriol. 1977 Dec;132(3):1027–1030. doi: 10.1128/jb.132.3.1027-1030.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Regulation of periodic protein synthesis in the cell cycle: control of initiation and termination of flagellar gene expression. Cell. 1981 Apr;24(1):49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Weissborn A., Steinmann H. M., Shapiro L. Characterization of the proteins of the Caulobacter crescentus flagellar filament. Peptide analysis and filament organization. J Biol Chem. 1982 Feb 25;257(4):2066–2074. [PubMed] [Google Scholar]