Abstract
We report here the molecular cloning of an ≈1-Mb region of recurrent amplification at 20q13.2 in breast cancer and other tumors and the delineation of a 260-kb common region of amplification. Analysis of the 1-Mb region produced evidence for five genes, ZNF217, ZNF218, and NABC1, PIC1L (PIC1-like), CYP24, and a pseudogene CRP (Cyclophillin Related Pseudogene). ZNF217 and NABC1 emerged as strong candidate oncogenes and were characterized in detail. NABC1 is predicted to encode a 585-aa protein of unknown function and is overexpressed in most but not all breast cancer cell lines in which it was amplified. ZNF217 is centrally located in the 260-kb common region of amplification, transcribed in multiple normal tissues, and overexpressed in all cell lines and tumors in which it is amplified and in two in which it is not. ZNF217 is predicted to encode alternately spliced, Kruppel-like transcription factors of 1,062 and 1,108 aa, each having a DNA-binding domain (eight C2H2 zinc fingers) and a proline-rich transcription activation domain.
Studies in which comparative genomic hybridization was used have revealed ≈20 regions of recurrent increased DNA sequence copy number in breast tumors (1–3). These regions are predicted to encode dominantly acting genes that may play a role in tumor progression or response to therapy. To date, three of these regions have been associated with established oncogenes: ERBB2 at 17q12, MYC at 8q24, and CCND1 and EMS1 at 11q13. In breast cancer, ERBB2 and CCND1/EMS1 amplification and overexpression are associated with decreased life expectancy (4, 5), whereas MYC amplification has been associated with lymph node involvement, advanced stage, and an increased rate of relapse (6, 7). Efforts are now underway in several laboratories to identify oncogenes in the other regions of amplification.
Amplification at 20q13 is particularly interesting because this aberration occurs in a variety of tumor types and is associated with aggressive tumor behavior. The initial comparative genomic hybridization study showed increased copy number involving 20q13 in 40% of breast cancer cell lines and 18% of primary breast tumors (2). Since then, other comparative genomic hybridization studies have revealed copy-number gains at 20q13 in greater than 25% of cancers of the ovary (8), colon (9), head and neck (10), brain (11), and pancreas (12). This region was analyzed at higher resolution in breast tumors and cell lines by using fluorescence in situ hybridization (FISH), and a 1.5-Mb-wide region of recurrent amplification containing the cosmid clone RMC20C001 was defined (13, 14). Interphase FISH with RMC20C001 revealed low level (>1.5-fold) and high level (>3-fold) amplification in 29% and 7% of breast cancers, respectively (15). High level amplification was associated with an aggressive tumor phenotype (15, 16).
Increased copy number of chromosome 20q in cultured cells also has been associated with phenotypes characteristic of progressing tumors, including immortalization and genomic instability. Specifically, increased copy number at 20q11-qter has been observed frequently in human uroepithelial cells (17) and keratinocytes (18) after transfection with HPV16 E7 or HPV16, respectively. In addition, increased copy number at 20q13.2 has been associated with p53-independent genomic instability in some HPV16 E7-transfected human uroepithelial cell lines (19).
These studies suggest that increased expression of one or more genes on 20q and especially at 20q13.2 contribute to the evolution of breast and other solid tumors. Several candidate genes, including AIB1 (20), BTAK (21), CAS (22), and TFAP2C (23), that may contribute to the various phenotypes associated with increased copy number involving 20q have been identified. However, we describe here a region of recurrent increased copy number at 20q13.2 that does not involve these genes. Extensive analysis of this region identified four previously unknown genes, one known gene, and a pseudogene. Two of the novel genes, designated ZNF217 and NABC1 (Novel Amplified in Breast Cancer-1), were amplified and overexpressed in multiple cell lines and tumors and were characterized extensively.
MATERIALS AND METHODS
Cell and Tissue Sources.
Karyotypically normal, finite lifespan human mammary epithelial cells 161RM, 48RM, and 184RM and the immortalized line 184B5 were obtained from M. Stampfer (Lawrence Berkeley National Laboratory) and cultured as described (24). Cell lines SKBR3, BT474, MDA-MB-436, HBL100, MDA-MB-435, HS758T, and MCF7 were obtained from American Type Culture Collection (ATCC) and cultured as suggested by ATCC. MDA10 was obtained from the National Cancer Institute. The cell line 600MPE was provided by H. Smith (California Pacific Medical Center, San Francisco) and cultured as described (25). Primary breast tumor samples were obtained from the San Francisco Bay Area Breast Cancer SPORE repository or from Tampere University.
Physical Map.
YAC clones from the Center d’Etudes du Polymorphisme Humain and Integrated Molecular Analysis of Gene Expression Consortium cDNAs were identified in the Whitehead Institute for Biomedical Research/Massachusetts Institute of Technology Center for Genomic Research web site (http://www.genome.wi.nit.edu) and obtained from Research Genetics (Huntsville, AL). P1 and Baterial Artificial Chromosome libraries were screened by using PCR with sequence tagged sites (STSs) from 20q13.2 as described (13) or as recommended by the supplier (Research Genetics or Genome Systems, St. Louis). All clones were mapped to normal metaphase cells by using FISH to confirm cytogenetic location. The position and physical relationship of each clone to its neighbors were determined and verified by many methods, including STS content mapping, Southern blot hybridization, fiber FISH, DNA fingerprinting, and genome sequence analysis.
Copy Number Analysis with Interphase FISH.
All samples were screened for amplification at 20q13.2 by using FISH with probes prepared from clones RMC20C001, RMC20B4095, and/or a pool of clones made up of RMC20P4030, RMC20P4040, RMC20P4038, RMC20P4039, RMC20P4041, and RMC20P4042. Samples found to be amplified by using one of these probes were analyzed more extensively by using probes prepared from clones listed in Table 1. Dual-color FISH was performed as reported (14) by using a test probe and a reference probe at 20p prepared from RMC20B235 containing D20S894. Fifty to 100 intact, nonoverlapping nuclei were scored for each probe. Nuclei with more than 20 signals were scored as >20 because of the difficulty of accurate signal enumeration at high copy number. Tumor-infiltrating leukocytes were excluded based on their small size. FISH to normal fibroblasts was performed to confirm that the hybridization efficiency for each probe was high.
Table 1.
Overlapping genomic clones across a ≈1-Mb region at 20q13.2
| Clone identification | Type | Size, kb | Library coordinate |
|---|---|---|---|
| RMC20B4166 | BAC | 130 | 226I4 |
| RMC20B4123 | BAC | 94 | 189C22 |
| RMC20P4067 | P1 | 74 | 91B2 |
| RMC20P4068 | P1 | 50 | 100D12 |
| RMC20P4042 | P1 | 57 | 103D9 |
| RMC20P4041 | P1 | 56 | 86C1 |
| RMC20B4095 | BAC | 80 | 140H15 |
| RMC20P4038 | P1 | 63 | 112G8 |
| RMC20P4039 | P1 | 74 | 34A6 |
| RMC20P4040 | P1 | 80 | 24H1 |
| RMC20P4030 | P1 | 102 | 124G6 |
| RMC20B4097 | BAC | 160 | 133E8 |
| RMC20P4007 | P1 | 56 | 31D11 |
| RMC20B4127 | BAC | 120 | 27P15 |
| RMC20B4103 | BAC | 77 | 188A15 |
| RMC20B4130 | BAC | 137 | 341H15 |
| RMC20P4185 | P1 | 83 | 1141D7 |
| RMC20B4188 | BAC | 103 | 163G21 |
| RMC20B4189 | BAC | 81 | 165E16 |
| RMC20P4002 | P1 | 83 | 12C6 |
| RMC20B4109 | BAC | 103 | 62A6 |
| RMC20B4108 | BAC | 48 | 432B9 |
| RMC20P4028 | P1 | 68 | 118G11 |
| RMC20P4010 | P1 | 75 | 36F10 |
| RMC20B4099 | BAC | 160 | 146L11 |
| RMC20P4018 | P1 | 64 | 77A10 |
| RMC20P4069 | P1 | 77 | 412B5 |
| RMC20B4122 | BAC | 108 | 39M7 |
| RMC20B4174 | BAC | 101 | 278I13 |
| RMC20B4087 | BAC | 87 | 96J14 |
Clones are listed in approximate genomic order from centromere to telomere. These clones are available from the Lawrence Berkeley National Laboratory/University of California, San Francisco Resource for Molecular Cytogenetics (RMC) upon request via (http://rmc-www.lbl.gov/).
Gene Fragment Identification.
A Lambda ZAP II (Stratagene) cDNA library constructed from the adenocarcinoma cell line Caco-2 (ATCC HTB 37) was screened with probes identified during direct cDNA selection and/or exon trapping as described (26). Selection of cDNAs by using the Genetrapper system was performed as recommended by the supplier (Life Technologies, Grand Island, NY). Exon trapping was accomplished as described (27). Direct selection of cDNA clones from a library generated from poly(A)+ RNA isolated from the cell line BT474 (ATCC HTB 20) was performed as described (28). Gene fragments selected by these methods were sequenced (SEQwright, Houston) and mapped back onto the clones from which they were selected by using PCR and/or Southern hybridization.
Northern Blot Analysis.
Total RNA from tumors or cell lines was extracted by Trizol separation as recommended by the supplier (Life Technologies). Each RNA sample (15 μg) was heated at 60°C for 15 min and loaded onto a 0.67 M formaldehyde/1.0× Mops/1.0% (wt/vol) agarose denaturing gel. After electrophoresis, RNA was transferred to a positively charged nylon membrane (Boehringer Mannheim) with 20× SSC and immobilized by UV crosslinking and baking at 70°C for 2 h. Hybridizations were carried out in Church buffer (29). All membranes were washed in 0.2× SSC/0.1% SDS at 60°C before detection. Hybridized probe was detected autoradiographically or by using a Molecular Dynamics PhosphorImager.
Quantitative PCR (QPCR).
Total RNA was isolated from cell lines and frozen tumor tissue by using Trizol separation. Frozen tissue was disrupted in Trizol by using a hand homogenizer before separation. RNA pellets were dissolved in deionized formamide to a concentration >1 μg/μl and stored at −20°C. Total RNA was treated with DNase I (Life Technologies) to remove any contaminating genomic DNA before reverse transcription. This RNA (15 ng) was reverse transcribed in a 100-μl reaction consisting of 1× PCR buffer II (Perkin–Elmer; 50 mM KCl/10 mM Tris, pH 8.3), 5.5 mM MgCl2, 500 μM each 2′-deoxynucleoside 5′-triphosphate, 2.5 μM random hexamers, 0.4 unit/μl RNase inhibitor, and 2.5 unit/μl Superscript II reverse transcriptase (Life Technologies). Reverse transcription was carried out at 25°C for 10 min, 48°C for 30 min, and 95°C for 5 min and then incubated at 4°C. As a control for genomic contamination, 15 ng of DNase-treated RNA was processed as described above except that reverse transcriptase was not added.
The relative abundances of ZNF217 and GAPDH mRNAs were assessed by using QPCR (30). A 105-bp fragment in exon 3 of ZNF217 was amplified by using primers: F1, 5′-GATGTTACTCCTCCTCCGGATG, and R1, 5′-CACACTTGGCCTGTATCTGCA. A TaqMan probe, 5′FAM (6-carboxy fluorescein)-AAAGAGAAGCAAACGGAGACCGCAGC-3′ TAMRA (6-carboxy-tetramethylrhodamine), was included during QPCR. PCR primers and the TaqMan probe for GAPDH were obtained from Perkin–Elmer. QPCR amplification was carried out in triplicate in 50-μl reaction volumes consisting of 1× PCR buffer A (Perkin–Elmer), 5.5 mM MgCl2, 0.4 μM each primer, 200 μM each 2′-deoxynucleoside 5′-triphosphate, 100 nM probe, and 0.025 unit/μl Taq Gold (Perkin–Elmer). cDNA (500 pg) or no reverse transcriptase control RNA was amplified according the thermal profile: 95°C for 10 min (95°C for 15 s, 60°C for 1 min) × 40 cycles and incubated at 25°C.
The number of cycles needed for FAM fluorescence from the ZNF217 and GAPDH reactions to cross a predetermined threshold was measured for each sample during QPCR by using an Applied Biosystems 7700 (31). The amount of starting ZNF217 mRNA relative to that for GAPDH for each sample, i, was calculated as 2ΔNi, where ΔNi was the difference in the number of cycles needed for the FAM fluorescence intensity for the ZNF217 and GAPDH reactions to reach a threshold value. All measurements were normalized to the ZNF217/GAPDH ratio for the cell line MDA10, which was analyzed in every experiment. Thus, the ZNF217/GAPDH relative expression level for each sample was REi = 2(ΔNi − ΔNcal), where ΔNcal was the difference in the number of cycles needed for the FAM fluorescence intensities from the ZNF217 and GAPDH reactions to reach a threshold value in the MDA10 reference.
Expression of Exons, cDNAs, and ESTs (Expressed Sequence Tags).
The presence of specific exons, cDNAs or ESTs in first-strand cDNA from the cell line BT474 or multiple cDNA libraries (Life Technologies) was determined by PCR amplification. Sequence-specific primers were designed by using MacVector software after screening for repetitive elements by using the Baylor College of Medicine (BCM) web server. Amplification reactions in 10-μl volumes were carried out with 1 μl of first-strand cDNA or 50 ng of library cDNA template by using capillary thermocyclers (Idaho Technologies, Idaho Falls, ID). The first-strand cDNA from BT474 was prepared from 2 μg of total RNA that had been digested with RNase-free DNase I. The RNA was incubated with 250 ng of oligo(dT)12–18 and 25 ng of random hexamers at 70°C for 10 min and then placed on ice. First-strand synthesis was carried out in 1× first-strand buffer, 0.5 mM each dATP, dCTP, dGTP, and dTTP, and 5 mM DTT (Life Technologies). After 1 h, the reaction was stopped by incubation at 55°C for 10 min. The remaining RNA was degraded by addition of RNase H, and the mixture was put at 55°C for an additional 10 min.
DNA Sequencing.
SEQwright performed all cDNA sequencing. Sequence manipulation and analysis was performed by using macvector (Oxford Molecular, Campbell, CA) and the BCM nucleic acid and protein search launchers (http://kiwi.imgen.bcm.tmc.edu:8088/search-launcher/launcher.html). All genome sequencing was performed in the Lawrence Berkeley National Laboratory Human Genome Center by using a directed sequencing strategy (32).
DNA and Protein Sequence Analysis.
DNA sequences and predicted proteins were searched against public nucleic acid and protein databases by using BLAST, BLASTX, TBLASTX, BLASTX+BEAUTY, BLASTP, and BLASTP + BEAUTY algorithms (33) via the BCM search launcher and macvector software. Repetitive elements were identified by using the BCM repeatmasker. Genomic sequence was analyzed by using xgrail (34), grail1a, sorfind (35), and genotator (36). Protein sequence was analyzed by using blocks and ppsearch via BCM and MacVector software.
RESULTS
Molecular cloning of a 1-Mb Region at 20q13.2.
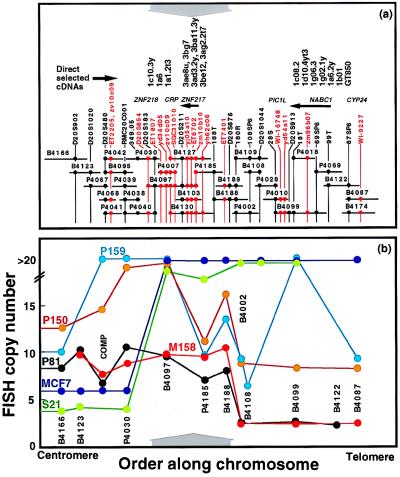
Overlapping P1 and BAC clones were assembled across a ≈1-Mb interval of 20q13.2 containing RMC20C001 to provide the framework for high-resolution analysis of DNA sequence copy number and for gene discovery. Thirty overlapping BAC and P1 clones and connecting STSs and ESTs are shown in Fig. 1a. The clone names, library coordinates, and sizes are listed in Table 1. The region extends from D20S902 to WI-9227 and is covered with Center d’Etudes du Polymorphisme Humain YACs 847g7, 845f3, and 820f5. A comprehensive description of the map and its genome sequence will be published elsewhere (C.C., unpublished data).
Figure 1.
(a) Physical map of 20q13.2 between D20S902 and WI-9227. Clones that make up the map are indicated as horizontal lines. The lengths of the lines are proportional to the sizes of the clones. The last five digits of the RMC identifiers listed in Table 1 identify clones. Mapped STSs and ESTs in this region are shown as vertical black and red lines, respectively. •, Clones that tested positive for STS or EST content by PCR. The approximate genome locations of five genes and a pseudogene are shown above the map. Arrowheads indicate transcriptional polarity where known. The approximate locations of 17 of 30 directly selected cDNA fragments are shown at the top. (b) FISH analysis of DNA sequence copy number in five informative breast tumors and one breast cancer cell line. Probes used in this study are indicated. The probe labeled COMP comprised clones P4030, P4040, P4038, P4039, P4041, and P4042. The vertical axis shows the number of hybridization signals produced by FISH with each probe. The number of signals is truncated at 20 because hybridization signal enumeration was difficult above this level. The x axis scales are the same for a and b. The results of six FISH studies are color coded by sample. Solid lines connect the measurements made for each sample. A ≈260-kb region of common maximal amplification is indicated by gray arrows above (a) and below (b).
DNA Sequence Copy Number Analysis.
Approximately 280 breast cancer tissue samples and cell lines were screened for increased DNA sequence copy number by using interphase FISH. Seventeen primary breast tumors and cell lines with >3× amplification relative to 20p were identified and analyzed for copy number at several points across the 1-Mb region by using FISH. In 11 of these, most of the 1-Mb region was amplified. However, the pattern of amplification in six suggested a ≈260-kb-wide region of maximal amplification extending from RMC20B4097 to RMC20B4188 (Fig. 1b). Copy number in the cell line MCF-7 and tumor S21 increased sharply at RMC20B4097 and extended distally, whereas in tumors P81 and M158, copy number increased sharply at RMC20B4188 and extended proximally. The copy number in tumors P150 and P159 also was high in the common region of maximal amplification. However, tumor P159 (Fig. 1b) and several others (not shown) had complex amplification patterns that hinted at the existence of other sites of amplification.
Gene Discovery.
Clones composing the ≈1-Mb contig were analyzed for gene content by using genomic sequencing (37), exon trapping (38), and direct cDNA selection (28) with cDNA prepared from the breast cancer cell line BT474. The region defined by RMC20P4185, RMC20B4188, and RMC20P4002 was analyzed twice by using exon trapping and cDNA selection because of the paucity of putative gene fragments that were recovered. Thirty-one trapped exons and 30 cDNAs were identified and sequenced. Whereas trapped exons were distributed broadly over the 1-Mb region, the cDNAs clustered as illustrated in Fig. 1a. ESTs either were identified by searching dBEST with sequence from genomic clones, trapped exons, and cDNA clones or were identified in the Whitehead and National Center for Biotechnology Information databases.
Several methods were used to determine the transcription patterns of the trapped exons, cDNA fragments, and ESTs. First-strand cDNA synthesized from total RNA isolated from the breast cancer cell line BT474 and DNA from multiple cDNA libraries were screened by using PCR with primer pairs designed from the DNA fragments. In addition, Northern blots of multiple tissue poly(A)+ mRNA samples and breast cancer cell line total RNA were probed. Independent DNA fragments that had the same approximate genome locations and patterns of transcription were assumed to be segments of one gene. These investigations provided evidence for three novel transcribed genes designated ZNF217, ZNF218, and NABC1. These were considered to be oncogene candidates and were characterized further (see below).
Evidence also was found for two other genes and a pseudogene in the 1-Mb region. These included CRP (Cyclophillin Related Pseudogene), CYP24 (vitamin D 24-hydroxylase), and PIC1L (PIC1-like). CRP was discovered in the genomic sequence by its significant DNA and amino acid alignment to cyclophilin (39). However, its ORF was disrupted by in-frame stop codons. CYP24 was previously reported to be involved in the regulation of calcium homeostasis (40). PIC1L was identified based on the homology of its predicted protein sequence to that for PIC1 (93% similarity), a nuclear body protein that interacts with PML in promyelocytic leukemia (41). Reverse transcription–PCR analyses of total mRNA from several breast cancer cell lines including BT474 revealed no evidence of expression for any of these sequences, and they were not analyzed further.
Transcribed Gene Descriptions.
ZNF218 was inferred from the transcription patterns detected by ET2205, direct selected cDNAs 1c10.3y, 1a6, and 1a1.2t3, and EST zv10c09. Hybridization of each of these DNA fragments to multitissue Northern blots revealed transcripts of ≈11, 8, and 7 kb in several tissues, including prostate, testis, ovary, and intestine, and a single transcript of 7 kb in the liver (data not shown). However, they did not detect transcription in breast cancer cell lines amplified at RMC20B4097. Interestingly, EST zv10c09 partially overlapped ET2205 and also mapped to a second site ≈200 kb distal to ET2205, suggesting that it is composed of two exons separated by a large intron.
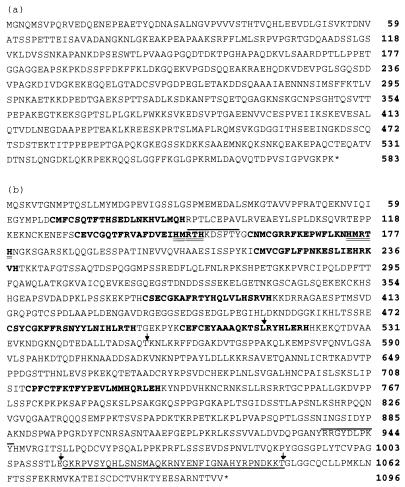
NABC1 was identified by seven direct selected cDNA fragments (1c08.2, 1d10.4yt3, 1g06.3, 1g02.1y, 1a6.2y, 1b01, and GT850; Fig. 1a). Hybridization of each these DNA fragments to multitissue Northern blots revealed a ≈3-kb transcript present at high abundance in brain and prostate and at lower abundance in testis, intestine, and colon (data not shown). Hybridization to Northern blots of total RNA from breast cancer cell lines and cultured epithelial cells derived from reduction mammoplasties showed the 3-kb transcript to be present at high levels in three of four cell lines amplified at RMC20B4099 but not detectable in normal epithelial cells (Fig. 2c). Northern hybridization to total RNA extracted from four primary breast tumors and the corresponding normal tissue revealed high level transcription in at least one tumor (Fig. 2e). However, NABC1 was not in the common region of maximal amplification, and transcription was not detected in the breast cancer cell line MCF7 in which it was amplified (Fig. 2c). A 2813-bp NABC1 cDNA was isolated from a colon carcinoma cDNA library and sequenced (accession no. AF041260). Analysis of the cDNA sequence identified a 1752-bp ORF predicted to encode a protein of 584 amino acids with no significant homology to other proteins (Fig. 3a).
Figure 2.
Expression of ZNF217 and NABC1. (a) Hybridization of a ZNF217 probe to multiple tissue poly(A)+ Northern blots. (b) Northern blot analysis of ZNF217 transcription in three independent karyotypically normal breast epithelial cultures of finite lifespan (161RM, 48RM, 185RM), one immortalized line (184BS) and five breast cancer cell lines (MDA10, SKBR3, MDA436, BT474, MCF7). The breast cancer cell lines carry 4, 10–15, 10–15, >20, and >20 copies of the region encoding ZNF217, respectively. (c) Northern blot analysis of NABC1 transcription in the same cell lines. (d) Analysis of ZNF217 transcription in four paired breast tumor/normal epithelium samples. Lanes containing RNA from normal and tumor tissues are labeled N and T, respectively. (e) Analysis of NABC1 transcription by using the same membrane described in d. (f) QPCR analysis of ZNF217 mRNA abundance relative to that for GAPDH in 11 primary breast tumors and nine breast cancer cell lines. DNA sequence copy number at the ZNF217 locus determined by FISH with RMC20B4097 is indicated above each QPCR measurement.
Figure 3.
Predicted amino acid sequences. (a) NABC1. (b) ZNF217. Eight putative C2H2 zinc finger domains are indicated in bold. A repeated motif in exon 1 is double underlined. Lines above the text indicate the positions of two putative tyrosine phosphorylation sites predicted by the PROSITE pattern search algorithm. A putative proline-rich transcription activation domain is encoded by amino acids 757-1005 and is composed of 16% proline. Exon boundaries were determined from the cDNA and genomic sequences and are marked with arrows. Exon 1 codes for 455 aa and exon 3 codes for 517 aa. The amino acid sequence of the alternately processed exon 4 is underlined.
ZNF217 was inferred from the pattern of transcription detected by exon ET6702, ESTs yj05a10 and zm10b10, and direct selected cDNAs 3ae8u, 3bg7, 3ad3.2y, 3ba11.3y, 3be12, and 3ag2.2t7. These DNA fragments mapped to the distal end of RMC20B4097 (Fig. 1a). They detected a major ≈6-kb transcript in most tissues except adult brain and fetal kidney. An additional ≈4-kb transcript was present in testis (Fig. 2a). The relative abundance of the major ZNF217 transcript was higher in breast cancer cell lines that were amplified at RMC204097 (MCF7, BT474, SKBR3, and MDA436) than in a nonamplified breast cancer cell line (MDA10) or cultured epithelial cells derived from reduction mammoplasties (161RM, 48RM, 184RM, and 184B5; Fig. 2b). ZNF217 was transcribed at high level in one of four primary tumors analyzed by Northern blot hybridization (Fig. 2d). ZNF217 expression was studied further by using the QPCR. Eleven primary tumors and nine breast cancer cell lines were analyzed. Six of the tumors and four of the cell lines carried >15 copies of the region defined by RMC20B4097. ZNF217 was transcribed at high level in all of these (Fig. 2f). Significantly, ZNF217 also was transcribed at high level in the breast cancer cell line 600MPE in which it was not amplified.
ZNF217 cDNA and Genomic Organization.
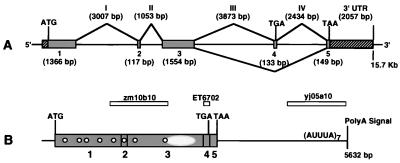
The genome and cDNA organization for ZNF217 shown in Fig. 4 were determined by sequencing directly selected cDNAs, trapped exons, ESTs, genome DNA, and cDNAs isolated from cDNA libraries. These analyses determined the complete 5,632-bp ZNF217 cDNA sequence (accession no. AF041259) and the 15.7-kb genome sequence in which it is encoded. Alignment of the cDNA and genome sequences revealed five coding exons flanked by canonical splice sites (Table 2). Analysis of the aligned consensus cDNA sequences identified alternately spliced ORFs of 3,186 and 3,324 bp with the structures indicated in Fig. 4. Alternative processing of the 133-bp exon 4 was suggested by the alignment of cDNAs from colon carcinoma and HeLa cell lines and exon ET6702.
Figure 4.
Genomic organization of ZNF217. (A) The genomic organization of the five exons with encoded initiation and termination codons that make up ZNF217. Exon 4 encodes a TGA termination codon and is alternatively processed. Hatched boxes represent known 5′- and 3′-untranslated regions (UTR) in the cDNA. The sizes of exons and introns appear below and above the map, respectively. (B) The map of the 5632-bp ZNF217 cDNA. Vertical bars represent exon boundaries. The relative positions of the predicted eight C2H2 Kruppel-like zinc finger motifs are indicated by white circles. The position of the proline-rich putative transcription activator domain is shown as a hatched oval. AUUUA motifs are indicated in the 3′-untranslated region. The relative locations of three ESTs are shown in boxes.
Table 2.
Sequence of exon boundaries in ZNF217
| No. | Exon, bp | Intron, bp | Exon-intron boundaries
|
|
|---|---|---|---|---|
| Splice acceptor junction | Splice donor junction | |||
| 1 | 1,366* | ATCCATCTGG/gtaagctgccct | ||
| 2 | 117 | 3,007 | ttttcctttaag/ATAAAAATGA | ACGCATACAG/gtaaagaacttt |
| 3 | 1,554 | 1,053 | ttcttgccttag/GTGAAAAACC | ACGTTAGAAG/gtattgcatgag |
| 4 | 133 | 3,873 | tttttcaatcag/GAAAAAGGCC | GGGAAAAAAG/gtgagcatatgt |
| 5 | 149* | 2,434 | ttttttccttag/GTCTTGGTGG | |
Exon sequences are shown in upper case; intron sequences are shown in lower case.
Translated portions.
Computational analysis of the translated ORFs indicated that ZNF217 encodes proteins of 1,062 and 1,108 aa (Fig. 3b). Both predicted proteins contained eight C2H2 zinc finger DNA-binding motifs. In addition, a proline-rich (16–20%) domain was located at residues 757–1,005 (Fig. 3b). Proline-rich domains have been shown to function as transcriptional activators in genes such as CTF/NF-1 (42). An imperfect polyadenylation signal (AATATA) was found 14 bases upstream of the poly(A) tail of a cDNA clone defined by EST yj05a10. Analysis of the ZNF217 3′-untranslated region revealed seven AUUUA motifs but no repetitive or conserved elements. The region of the mRNA sequence from nucleotide 5,211 to nucleotide 5,567 was composed of 44% uridine and contained five of the seven AUUUA motifs.
DISCUSSION
Several studies have suggested that amplification of genes on 20q contribute to neoplastic transformation and/or progression in breast cancer and other solid tumors. These include the association of increased copy number at 20q13 with high tumor grade and an aggressive tumor phenotype (14–16, 43) and the association between increased copy number at 20q and immortalization and genome instability in HPV-16-transfected cells (17–19). Several candidate oncogenes have been reported as amplified on 20q, but none of these are in the 1-Mb region of amplification analyzed in this study. AIB1 (20) and CAS (22) map proximal to this 1-Mb region at 20q12, and 20q13.1, respectively. TFAP2C (23) maps distal to the region by dual-color FISH (data not shown), and BTAK (21) maps distal to the 1-Mb region in Center d’Etudes du Polymorphisme Humain YAC 757e8.
Four previously unknown genes, PIC1L, ZNF217, ZNF218, and NABC1, and the known gene CYP24 were located in the 1-Mb region analyzed in this study. Three of these, CYP24, ZNF218, and PIC1L were excluded as candidate breast cancer genes because they were not expressed in breast cancer cell lines amplified at 20q13.2. NABC1 was highly expressed in three amplified breast cancer cell lines (SKBR3, BT474, and MDA436) and in one tumor without amplification at RMC20B4099. However, it was not expressed in MCF7 in which it is highly amplified (Fig. 1b). Furthermore, it mapped outside the 260-kb region of common maximal amplification defined in this study. Nevertheless, it remains a tantalizing candidate and merits further characterization.
ZNF217 has several features that suggest it is an oncogene involved in breast cancer. First, it is located in a narrowly defined region of recurrent maximal amplification apparently devoid of other transcribed genes. Moreover, DNA sequence analysis suggests that ZNF217 encodes a transcription factor with 8 C2H2 Kruppel-like (44) DNA-binding motifs and a proline-rich transcription activator domain. Kruppel-like transcription factors have been implicated in several human malignancies, including EVI1, PLZF, and PLM in leukemia, BCL6 in lymphomas, GLI1 in glioblastoma, WT1 in Wilms tumor, and ZBP89 in gastric carcinoma. The presence of mRNA-destabilizing AU-rich elements in its 3′-untranslated region further suggests that ZNF217 is an oncogene because these motifs have been implicated in the destabilization of mRNA transcripts of protooncogenes and nuclear transcription factors (e.g., FOS, JUN, MYC) and cytokines (GM-CSF, IL-3, b-IFN) (45). Finally, ZNF217 was transcribed at high levels in 10 of 10 tumors and cell lines with 20q13.2 amplification and 2 without amplification (cell line 600MPE and primary tumor T4) and at low levels in 17 of 19 tumors and cell lines without amplification. Thus, although amplification appears to be the predominant mechanism leading to overexpression, transcript abundance also may be increased by other mechanisms as occur for established oncogenes such as ERBB2, MYC, and NMYC.
Acknowledgments
The authors thank Dr. H.-L. Weier for assistance with map assembly by using fiber FISH and Dr. R. Jensen for help with the application of QPCR to analysis of relative gene expression. This work was supported by Breast Cancer SPORE Grant CA 58207, the U.S. Department of Energy under Contract DE-AC-03-76SF00098, and Vysis, Inc.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: FISH, fluorescence in situ hybridization; STS, sequence-tagged site; QPCR, quantitative PCR; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; EST, expressed sequence tag; BCM, Baylor College of Medicine.
References
- 1.Muleris M, Almeida A, Gerbault-Seureau M, Malfoy B, Dutrillaux B. Genes Chromosomes Cancer. 1994;10:160–170. doi: 10.1002/gcc.2870100303. [DOI] [PubMed] [Google Scholar]
- 2.Kallioniemi A, Kallioniemi O P, Piper J, Tanner M, Stokke T, Chen L, Smith H S, Pinkel D, Gray J W, Waldman F M. Proc Natl Acad Sci USA. 1994;91:2156–2160. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Isola J J, Kallioniemi O P, Chu L W, Fuqua S A, Hilsenbeck S G, Osborne C K, Waldman F M. Am J Pathol. 1995;147:905–911. [PMC free article] [PubMed] [Google Scholar]
- 4.Gaudray P, Szepetowski P, Escot C, Birnbaum D, Theillet C. Mutat Res. 1992;276:317–328. doi: 10.1016/0165-1110(92)90018-5. [DOI] [PubMed] [Google Scholar]
- 5.Borg A, Baldetorp B, Ferno M, Olsson H, Ryden S, Sigurdsson H. Oncogene. 1991;6:137–143. [PubMed] [Google Scholar]
- 6.Borg A, Baldetorp B, Ferno M, Olsson H, Sigurdsson H. Int J Cancer. 1992;51:687–691. doi: 10.1002/ijc.2910510504. [DOI] [PubMed] [Google Scholar]
- 7.Berns E M, Foekens J A, van Staveren I L, van Putten W L, de Koning H Y, Portengen H, Klijn J G. Gene. 1995;159:11–18. doi: 10.1016/0378-1119(94)00534-y. [DOI] [PubMed] [Google Scholar]
- 8.Iwabuchi H, Sakamoto M, Sakunaga H, Yen-Ming M, Carcanyin M L, Pinkel D, Yang-Feng T L, Gray J W. Cancer Res. 1995;55:6172–6180. [PubMed] [Google Scholar]
- 9.Schlegel J, Stumm G, Scherthan H, Bocker T, Zirngibl H, Ruschoff J, Hofstadter F. Cancer Res. 1995;55:6002–6005. [PubMed] [Google Scholar]
- 10.Bockmuhl U, Petersen I, Schwendel A, Dietel M. Laryngorhinootologie. 1996;75:408–414. doi: 10.1055/s-2007-997605. [DOI] [PubMed] [Google Scholar]
- 11.Mohapatra G, Kim D H, Feuerstein B G. Genes Chromosomes Cancer. 1995;13:86–93. doi: 10.1002/gcc.2870130203. [DOI] [PubMed] [Google Scholar]
- 12.Solinas-Toldo S, Wallrapp C, Muller-Pillasch F, Bentz M, Gress T, Lichter P. Cancer Res. 1996;56:3803–3807. [PubMed] [Google Scholar]
- 13.Stokke T, Collins C, Kuo W L, Kowbel D, Shadravan F, Tanner M, Kallioniemi A, Kallioniemi O P, Pinkel D, Deaven L, et al. Genomics. 1995;26:134–137. doi: 10.1016/0888-7543(95)80092-z. [DOI] [PubMed] [Google Scholar]
- 14.Tanner M M, Tirkkonen M, Kallioniemi A, Collins C, Stokke T, Karhu R, Kowbel D, Shadravan F, Hintz M, Kuo W L, et al. Cancer Res. 1994;54:4257–4260. [PubMed] [Google Scholar]
- 15.Tanner M M, Tirkkonen M, Kallioniemi A, Holli K, Collins C, Kowbel D, Gray J W, Kalliomiemi O P, Isola J. Clin Cancer Res. 1995;1:1455–1461. [PubMed] [Google Scholar]
- 16.Courjal F, Cuny M, Rodriguez C, Louason G, Speiser P, Katsaros D, Tanner M M, Zeillinger R, Theillet C. Br J Cancer. 1996;74:1984–1989. doi: 10.1038/bjc.1996.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reznikoff C A, Belair C, Savelieva E, Zhai Y, Pfeifer K, Yeager T, Thompson K J, DeVries S, Bindley C, Newton M A, et al. Genes Dev. 1994;8:2227–2240. doi: 10.1101/gad.8.18.2227. [DOI] [PubMed] [Google Scholar]
- 18.Solinas-Toldo S, Durst M, Lichter P. Proc Natl Acad Sci USA. 1997;94:3854–3859. doi: 10.1073/pnas.94.8.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savelieva E, Belair C D, Newton M A, DeVries S, Gray J W, Waldman F, Reznikoff C A. Oncogene. 1997;14:551–560. doi: 10.1038/sj.onc.1200868. [DOI] [PubMed] [Google Scholar]
- 20.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 21.Sen S, Zhou H, White R A. Oncogene. 1997;14:2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 22.Brinkmann U, Gallo M, Polymeropoulos M H, Pastan I. Genome Res. 1996;6:187–194. doi: 10.1101/gr.6.3.187. [DOI] [PubMed] [Google Scholar]
- 23.Williamson J A, Bosher J M, Skinner A, Sheer D, Williams T, Hurst H C. Genomics. 1996;35:262–264. doi: 10.1006/geno.1996.0351. [DOI] [PubMed] [Google Scholar]
- 24.Stampfer M R, Yaswen P. Cancer Surv. 1993;18:7–34. [PubMed] [Google Scholar]
- 25.Smith H, Wolman S, Dairkee S, Hancock M, Lippman M, Leff A, Hackett A. Natl Cancer Inst Monogr. 1987;78:611–615. [PubMed] [Google Scholar]
- 26.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning. A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 27.Church D M, Stotler C J, Rutter J L, Murrell J R, Trofatter J A, Buckler A J. Nat Genet. 1994;6:98–105. doi: 10.1038/ng0194-98. [DOI] [PubMed] [Google Scholar]
- 28.Rommens J M, Mar L, McArthur J, Tsui L-C, Scherer S W. In: Toward a Transcriptional Map of the q21–q22 Region of Chromosome 7. Hochgeschwender U, Gardiner K, editors. New York: Plenum; 1994. [Google Scholar]
- 29.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson U E, Heid C A, Williams P M. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 31.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 32.Kimmerly W J, Kyle A L, Lustre V M, Martin C H, Palazzolo M J. Genet Anal Tech Appl. 1994;11:117–128. doi: 10.1016/1050-3862(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 33.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Uberbacher E C, Xu Y, Mural R J. Methods Enzymol. 1996;266:259–281. doi: 10.1016/s0076-6879(96)66018-2. [DOI] [PubMed] [Google Scholar]
- 35.Hutchinson G B, Hayden M R. Nucleic Acids Res. 1992;20:3453–3462. doi: 10.1093/nar/20.13.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris N L. Genome Res. 1997;7:754–762. doi: 10.1101/gr.7.7.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin C H, Mayeda C A, Davis C A, Ericsson C L, Knafels J D, Mathog D R, Celniker S E, Lewis E B, Palazzolo M J. Proc Natl Acad Sci USA. 1995;92:8398–8402. doi: 10.1073/pnas.92.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buckler A J, Chang D D, Graw S L, Brook J D, Haber D A, Sharp P A, Housman D E. Proc Natl Acad Sci USA. 1991;88:4005–4009. doi: 10.1073/pnas.88.9.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haendler B, Keller R, Hiestand P C, Kocher H P, Wegmann G, Movva N R. Gene. 1989;83:39–46. doi: 10.1016/0378-1119(89)90401-0. [DOI] [PubMed] [Google Scholar]
- 40.Ohyama Y, Noshiro M, Okuda K. FEBS Lett. 1991;278:195–198. doi: 10.1016/0014-5793(91)80115-j. [DOI] [PubMed] [Google Scholar]
- 41.Boddy M N, Howe K, Etkin L D, Solomon E, Freemont P S. Oncogene. 1996;13:971–982. [PubMed] [Google Scholar]
- 42.Mermod N, O’Neill E A, Kelly T J, Tjian R. Cell. 1989;58:741–753. doi: 10.1016/0092-8674(89)90108-6. [DOI] [PubMed] [Google Scholar]
- 43.Tanner M M, Tirkkonen M, Kallioniemi A, Isola J, Kuukasjarvi T, Collins C, Kowbel D, Guan X Y, Trent J, Gray J W, et al. Cancer Res. 1996;56:3441–3445. [PubMed] [Google Scholar]
- 44.Pieler T, Bellefroid E. Mol Biol Rep. 1994;20:1–8. doi: 10.1007/BF00999848. [DOI] [PubMed] [Google Scholar]
- 45.Chen C Y, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]