Abstract
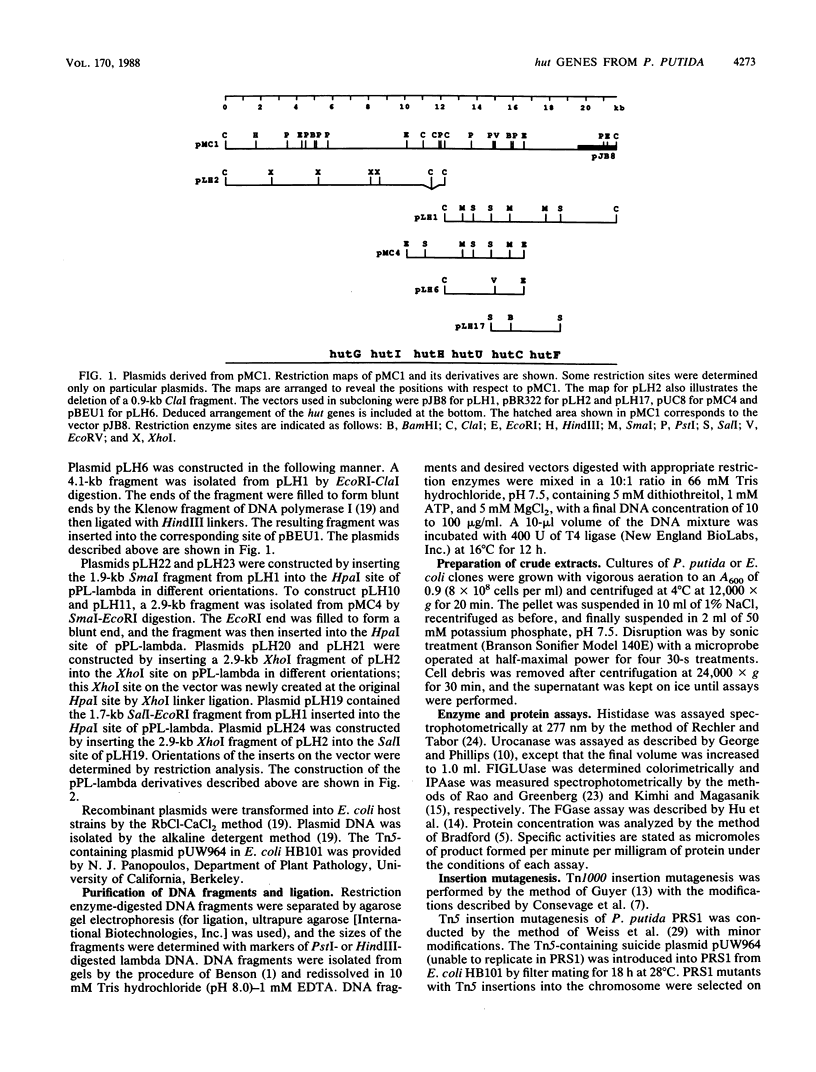
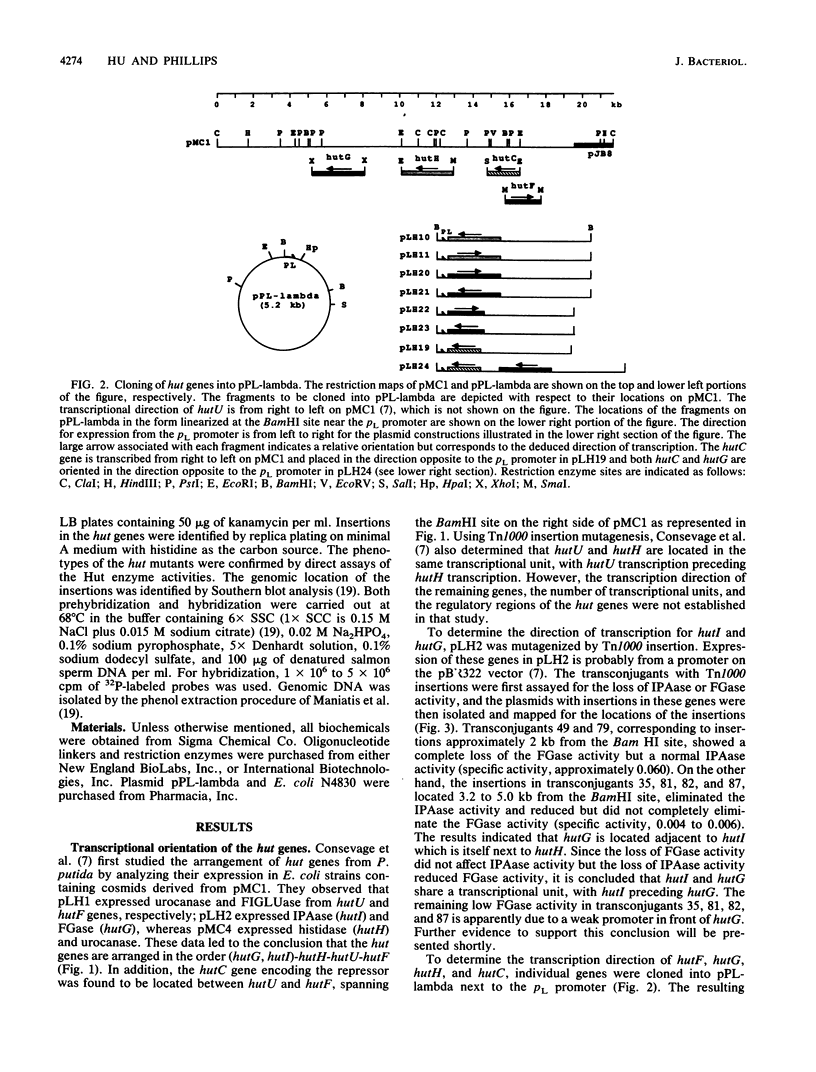
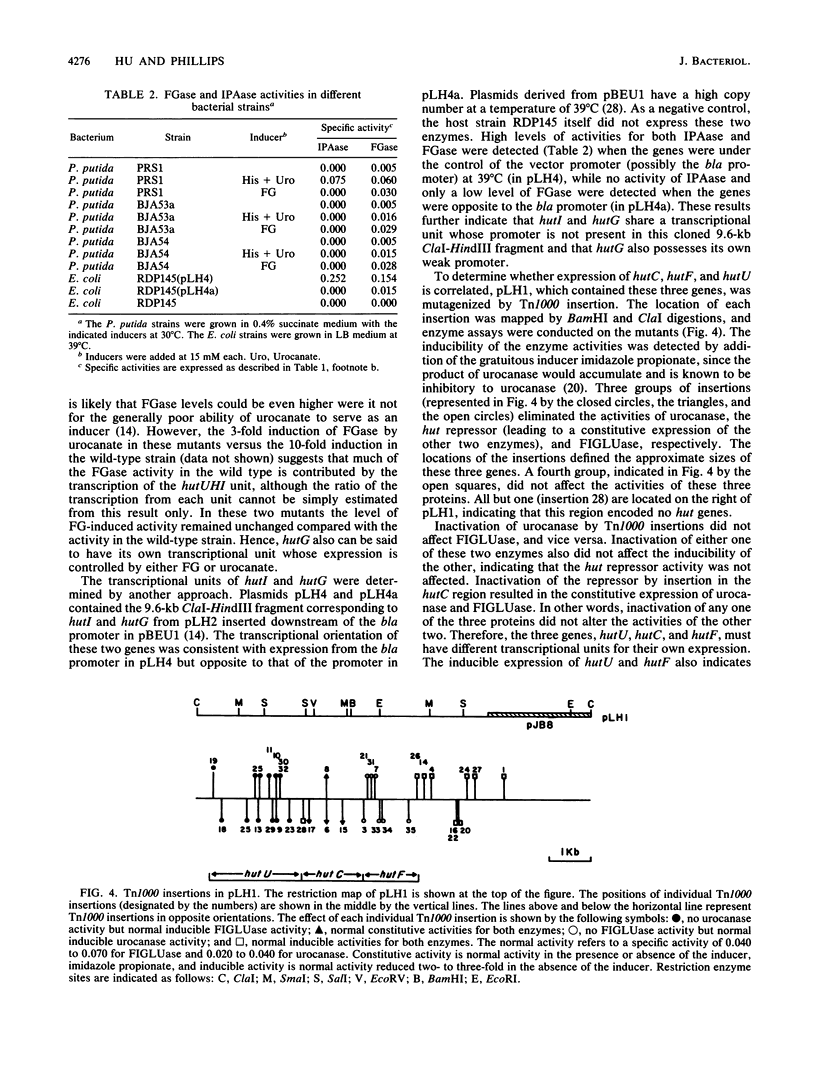
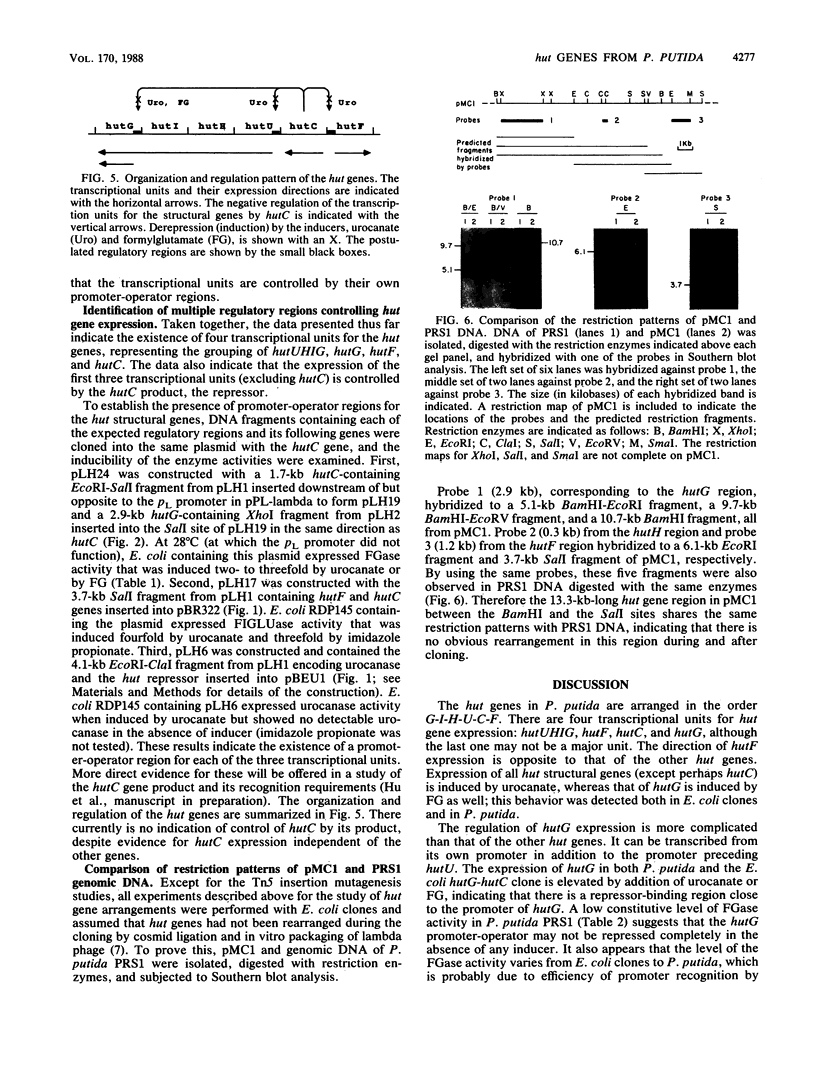
The arrangement of the histidine utilization (hut) genes in Pseudomonas putida was established by examining the structure of a DNA segment that had been cloned into Escherichia coli via a cosmid vector. Southern blot analysis revealed that the restriction patterns of the hut genes cloned into E. coli and present in the P. putida genome were identical, indicating that no detectable DNA rearrangement took place during the cloning. Expression of the hut genes from a series of overlapping clones indicated the gene order to be hutG-hutI-hutH-hutU-hutC-hutF. The transcription directions of the different hut genes were determined by cloning the genes under control of the lambda pL promoter. This showed that hutF, encoding formiminoglutamate hydrolase, was transcribed in a direction opposite to that of the other genes. Inactivation of the cloned hut genes by Tn1000 insertion revealed that the hut genes were divided into three major transcriptional units (hutF, hutC [the repressor gene], and hut UHIG), but hutG may also be independently transcribed. When cloned individually with hutC on the same vector, hutF and hutU (which encodes urocanase) expression was induced by urocanate, indicating that these two genes each possess an operator-promoter element. Tn1000 insertions (in the cloned genes) or Tn5 insertions (in the P. putida genome) affecting the hutI or hutH gene only partially eliminated hutG expression. Furthermore, hutG, which specifies N-formylglutamate amidohydrolase, was regulated by the hutC product when the two genes were cloned on the same vector and expressed in E. coli. Therefore, hutG can be expressed independently from its own promoter, in keeping with earlier observations that N-formylglutamate amidohydrolase synthesis is not coordinated with that of urocanase and histidase and can be induced by N-formylglutamate or urocanate.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blumenberg M., Magasanik B. Physical maps of Klebsiella aerogenes and Salmonella typhimurium hut genes. J Bacteriol. 1981 Jan;145(1):664–667. doi: 10.1128/jb.145.1.664-667.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Bender R. A. Genetic and physical maps of Klebsiella aerogenes genes for histidine utilization (hut). Mol Gen Genet. 1984;193(1):99–103. doi: 10.1007/BF00327421. [DOI] [PubMed] [Google Scholar]
- Boylan S. A., Eades L. J., Janssen K. A., Lomax M. I., Bender R. A. A restriction enzyme cleavage map of the histidine utilization (hut) genes of Klebsiella aerogenes and deletions lacking regions of hut DNA. Mol Gen Genet. 1984;193(1):92–98. doi: 10.1007/BF00327420. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Consevage M. W., Porter R. D., Phillips A. T. Cloning and expression in Escherichia coli of histidine utilization genes from Pseudomonas putida. J Bacteriol. 1985 Apr;162(1):138–146. doi: 10.1128/jb.162.1.138-146.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote J. G., Hassall H. The control of the enzymes degrading histidine and related imidazolyl derivates in Pseudomonas testosteroni. Biochem J. 1973 Mar;132(3):423–433. doi: 10.1042/bj1320423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coote J. G., Hassall H. The degradation of L-histidine, imidazolyl-L-lactate and imidazolylpropionate by Pseudomonas testosteroni. Biochem J. 1973 Mar;132(3):409–422. doi: 10.1042/bj1320409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George D. J., Phillips A. T. Identification of alpha-ketobutyrate as the prosthetic group of urocanase from Pseudomonas putida. J Biol Chem. 1970 Feb 10;245(3):528–537. [PubMed] [Google Scholar]
- Goldberg R. B., Bloom F. R., Magasanik B. Regulation of histidase synthesis in intergeneric hybrids of enteric bacteria. J Bacteriol. 1976 Jul;127(1):114–119. doi: 10.1128/jb.127.1.114-119.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg R. B., Magasanik B. Gene order of the histidine utilization (hut) operons in Klebsiella aerogenes. J Bacteriol. 1975 Jun;122(3):1025–1031. doi: 10.1128/jb.122.3.1025-1031.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Hu L., Mulfinger L. M., Phillips A. T. Purification and properties of formylglutamate amidohydrolase from Pseudomonas putida. J Bacteriol. 1987 Oct;169(10):4696–4702. doi: 10.1128/jb.169.10.4696-4702.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimhi Y., Magasanik B. Genetic basis of histidine degradation in Bacillus subtilis. J Biol Chem. 1970 Jul 25;245(14):3545–3548. [PubMed] [Google Scholar]
- Lessie T. G., Neidhardt F. C. Formation and operation of the histidine-degrading pathway in Pseudomonas aeruginosa. J Bacteriol. 1967 Jun;93(6):1800–1810. doi: 10.1128/jb.93.6.1800-1810.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P., Magasanik B. N-formimino-L-glutamate formiminohydrolase of Aerobacter aerogenes. J Biol Chem. 1965 Nov;240(11):4316–4319. [PubMed] [Google Scholar]
- Matherly L. H., Phillips A. T. Substrate-mediated inactivation of urocanase from Pseudomonas putida. Evidence for an essential sulfhydryl group. Biochemistry. 1980 Dec 9;19(25):5814–5818. doi: 10.1021/bi00566a024. [DOI] [PubMed] [Google Scholar]
- Parada J. L., Magasanik B. Expression of the hut operons of Salmonella typhimurium in Klebsiella aerogenes and in Escherichia coli. J Bacteriol. 1975 Dec;124(3):1263–1268. doi: 10.1128/jb.124.3.1263-1268.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAO D. R., GREENBERG D. M. Studies on the enzymic decomposition of urocanic acid. IV. Purification and properties of 4(5)-imidazolone-5(4)-propionic acid hydrolase. J Biol Chem. 1961 Jun;236:1758–1763. [PubMed] [Google Scholar]
- Smith G. R., Magasanik B. The two operons of the histidine utilization system in Salmonella typhimurium. J Biol Chem. 1971 May 25;246(10):3330–3341. [PubMed] [Google Scholar]
- Tyler B. M., Goldberg R. B. Transduction of chromosomal genes between enteric bacteria by bacteriophage P1. J Bacteriol. 1976 Mar;125(3):1105–1111. doi: 10.1128/jb.125.3.1105-1111.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlin B. E., Clark A. J. Overproduction of the Escherichia coli recA protein without stimulation of its proteolytic activity. J Bacteriol. 1981 Oct;148(1):386–390. doi: 10.1128/jb.148.1.386-390.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss A. A., Hewlett E. L., Myers G. A., Falkow S. Tn5-induced mutations affecting virulence factors of Bordetella pertussis. Infect Immun. 1983 Oct;42(1):33–41. doi: 10.1128/iai.42.1.33-41.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]