Abstract
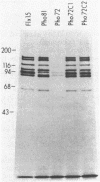
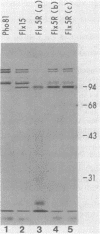
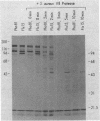
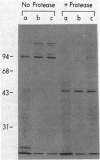
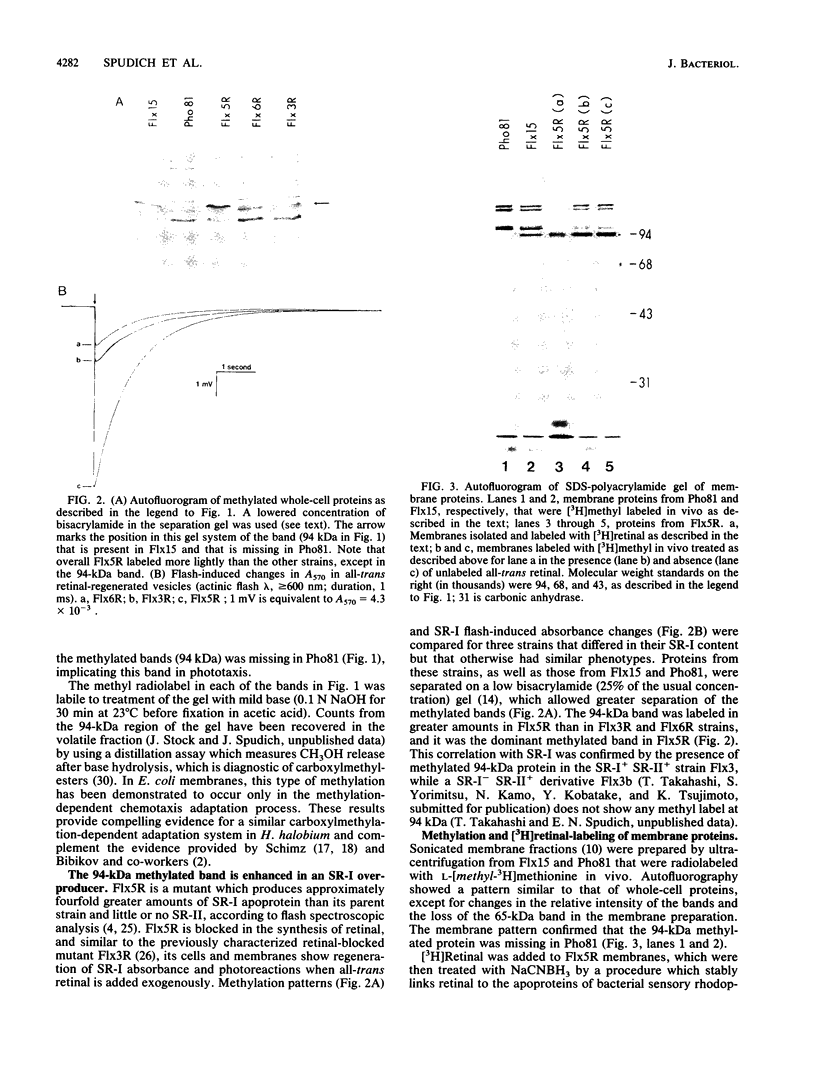
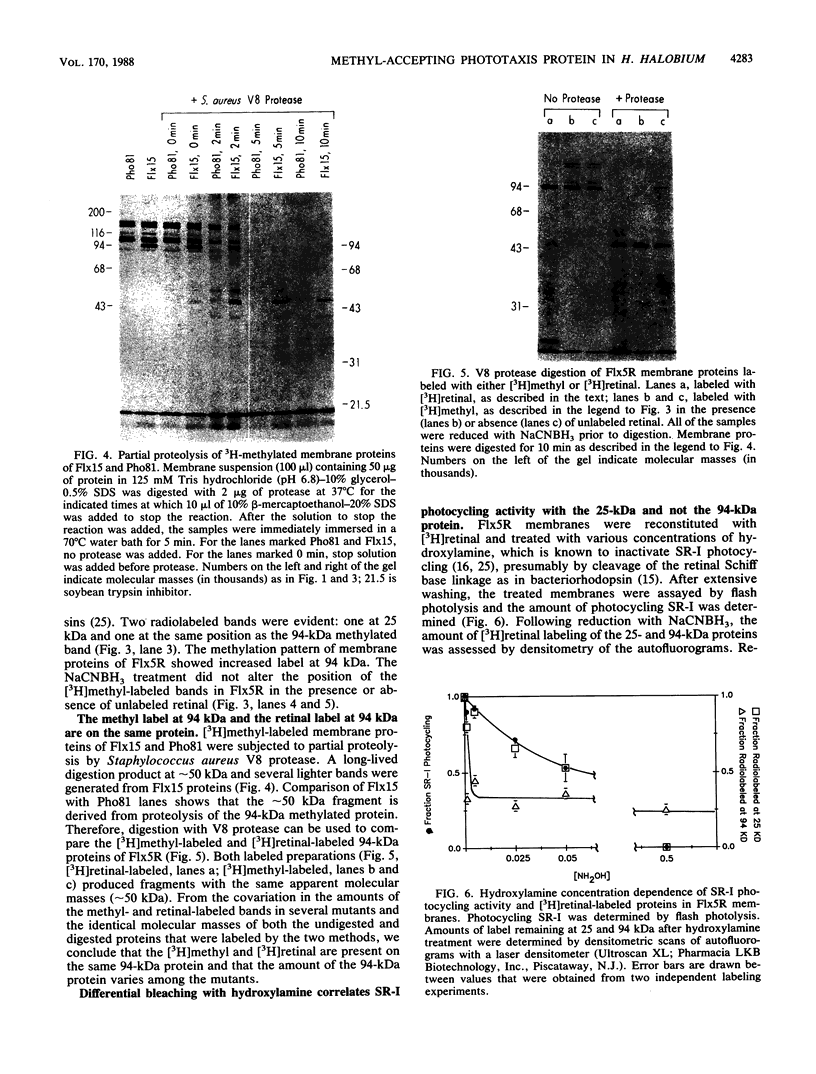
In vivo radiolabeling of Halobacterium halobium phototaxis mutants and revertants with L-[methyl-3H] methionine implicated seven methyl-accepting protein bands with apparent molecular masses from 65 to 150 kilodaltons (kDa) in adaptation of the organism to chemo and photo stimuli, and one of these (94 kDa) was specifically implicated in phototaxis. The lability of the radiolabeled bands to mild base treatment indicated that the methyl linkages are carboxylmethylesters, as is the case in the eubacterial chemotaxis receptor-transducers. The 94-kDa protein was present in increased amounts in an overproducer of the apoprotein of sensory rhodopsin I, one of two retinal-containing phototaxis receptors in H. halobium. It was absent in a strain that contained sensory rhodopsin II and that lacked sensory rhodopsin I and was also absent in a mutant that lacked both photoreceptors. Based on the role of methyl-accepting proteins in chemotaxis in other bacteria, we suggest that the 94-kDa protein is the signal transducer for sensory rhodopsin I. By [3H]retinal labeling studies, we previously identified a 25-kDa retinal-binding polypeptide that was derived from photochemically reactive sensory rhodopsin I. When H. halobium membranes containing sensory rhodopsin I were treated by a procedure that stably reduced [3H]retinal onto the 25-kDa apoprotein, a 94-kDa protein was also found to be radiolabeled. Protease digestion confirmed that the 94-kDa retinal-labeled protein was the same as the methyl-accepting protein that was suggested above to be the signal transducer for sensory rhodopsin I. Possible models are that the 25- and 94-kDa proteins are tightly interacting components of the photosensory signaling machinery or that both are forms of sensory rhodopsin I.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogomolni R. A., Spudich J. L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolni R. A., Spudich J. L. The photochemical reactions of bacterial sensory rhodopsin-I. Flash photolysis study in the one microsecond to eight second time window. Biophys J. 1987 Dec;52(6):1071–1075. doi: 10.1016/S0006-3495(87)83301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dencher N. A., Hildebrand E. Sensory transduction in Halobacterium halobium: retinal protein pigment controls UV-induced behavioral response. Z Naturforsch C. 1979 Sep-Oct;34(9-10):841–847. doi: 10.1515/znc-1979-9-1030. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Harayama S. Sensory transduction in bacterial chemotaxis. Int Rev Cytol. 1983;81:33–70. doi: 10.1016/s0074-7696(08)62334-7. [DOI] [PubMed] [Google Scholar]
- Katre N. V., Wolber P. K., Stoeckenius W., Stroud R. M. Attachment site(s) of retinal in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4068–4072. doi: 10.1073/pnas.78.7.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwai O., Hayashi H. Studies on bacterial chemotaxis. IV. Interaction of maltose receptor with a membrane-bound chemosensing component. J Biochem. 1979 Jul;86(1):27–34. [PubMed] [Google Scholar]
- Koshland D. E., Jr Biochemistry of sensing and adaptation in a simple bacterial system. Annu Rev Biochem. 1981;50:765–782. doi: 10.1146/annurev.bi.50.070181.004001. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K., MacDonald R. E. Light-induced transport in Halobacterium halobium. Methods Enzymol. 1979;56:398–407. doi: 10.1016/0076-6879(79)56038-8. [DOI] [PubMed] [Google Scholar]
- Marwan W., Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein (P480). J Mol Biol. 1987 May 20;195(2):333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- McCain D. A., Amici L. A., Spudich J. L. Kinetically resolved states of the Halobacterium halobium flagellar motor switch and modulation of the switch by sensory rhodopsin I. J Bacteriol. 1987 Oct;169(10):4750–4758. doi: 10.1128/jb.169.10.4750-4758.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlin D. M., Nettleton D. O., Ordal G. W., Hazelbauer G. L. Chemotactic transducer proteins of Escherichia coli exhibit homology with methyl-accepting proteins from distantly related bacteria. J Bacteriol. 1985 Jul;163(1):262–266. doi: 10.1128/jb.163.1.262-266.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D., Schuhmann L., Gruber H. Light-dependent reaction of bacteriorhodopsin with hydroxylamine in cell suspensions of Halobacterium halobium: demonstration of an apo-membrane. FEBS Lett. 1974 Aug 30;44(3):257–261. doi: 10.1016/0014-5793(74)81152-x. [DOI] [PubMed] [Google Scholar]
- Scherrer P., McGinnis K., Bogomolni R. A. Biochemical and spectroscopic characterization of the blue-green photoreceptor in Halobacterium halobium. Proc Natl Acad Sci U S A. 1987 Jan;84(2):402–406. doi: 10.1073/pnas.84.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimz A., Hildebrand E. Chemosensory responses of Halobacterium halobium. J Bacteriol. 1979 Dec;140(3):749–753. doi: 10.1128/jb.140.3.749-753.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimz A. Methylation of membrane proteins is involved in chemosensory and photosensory behavior of Halobacterium halobium. FEBS Lett. 1981 Mar 23;125(2):205–207. doi: 10.1016/0014-5793(81)80719-3. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979 Jul 26;280(5720):279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Bogomolni R. A., Spudich J. L. Genetic and biochemical resolution of the chromophoric polypeptide of halorhodopsin. Biochem Biophys Res Commun. 1983 Apr 15;112(1):332–338. doi: 10.1016/0006-291x(83)91835-1. [DOI] [PubMed] [Google Scholar]
- Spudich E. N., Spudich J. L. Control of transmembrane ion fluxes to select halorhodopsin-deficient and other energy-transduction mutants of Halobacterium halobium. Proc Natl Acad Sci U S A. 1982 Jul;79(14):4308–4312. doi: 10.1073/pnas.79.14.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich E. N., Sundberg S. A., Manor D., Spudich J. L. Properties of a second sensory receptor protein in Halobacterium halobium phototaxis. Proteins. 1986 Nov;1(3):239–246. doi: 10.1002/prot.340010306. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984 Dec 6;312(5994):509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Sensory rhodopsins of halobacteria. Annu Rev Biophys Biophys Chem. 1988;17:193–215. doi: 10.1146/annurev.bb.17.060188.001205. [DOI] [PubMed] [Google Scholar]
- Spudich J. L., Bogomolni R. A. Spectroscopic discrimination of the three rhodopsinlike pigments in Halobacterium halobium membranes. Biophys J. 1983 Aug;43(2):243–246. doi: 10.1016/S0006-3495(83)84345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Clarke S., Koshland D. E., Jr The protein carboxylmethyltransferase involved in Escherichia coli and Salmonella typhimurium chemotaxis. Methods Enzymol. 1984;106:310–321. doi: 10.1016/0076-6879(84)06031-6. [DOI] [PubMed] [Google Scholar]
- Sundberg S. A., Alam M., Spudich J. L. Excitation signal processing times in Halobacterium halobium phototaxis. Biophys J. 1986 Nov;50(5):895–900. doi: 10.1016/S0006-3495(86)83530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg S. A., Bogomolni R. A., Spudich J. L. Selection and properties of phototaxis-deficient mutants of Halobacterium halobium. J Bacteriol. 1985 Oct;164(1):282–287. doi: 10.1128/jb.164.1.282-287.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T., Mochizuki Y., Kamo N., Kobatake Y. Evidence that the long-lifetime photointermediate of s-rhodopsin is a receptor for negative phototaxis in Halobacterium halobium. Biochem Biophys Res Commun. 1985 Feb 28;127(1):99–105. doi: 10.1016/s0006-291x(85)80131-5. [DOI] [PubMed] [Google Scholar]
- Tomioka H., Takahashi T., Kamo N., Kobatake Y. Flash spectrophotometric identification of a fourth rhodopsin-like pigment in Halobacterium halobium. Biochem Biophys Res Commun. 1986 Sep 14;139(2):389–395. doi: 10.1016/s0006-291x(86)80003-1. [DOI] [PubMed] [Google Scholar]
- Wolff E. K., Bogomolni R. A., Scherrer P., Hess B., Stoeckenius W. Color discrimination in halobacteria: spectroscopic characterization of a second sensory receptor covering the blue-green region of the spectrum. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7272–7276. doi: 10.1073/pnas.83.19.7272. [DOI] [PMC free article] [PubMed] [Google Scholar]