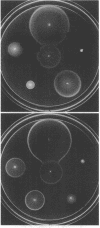
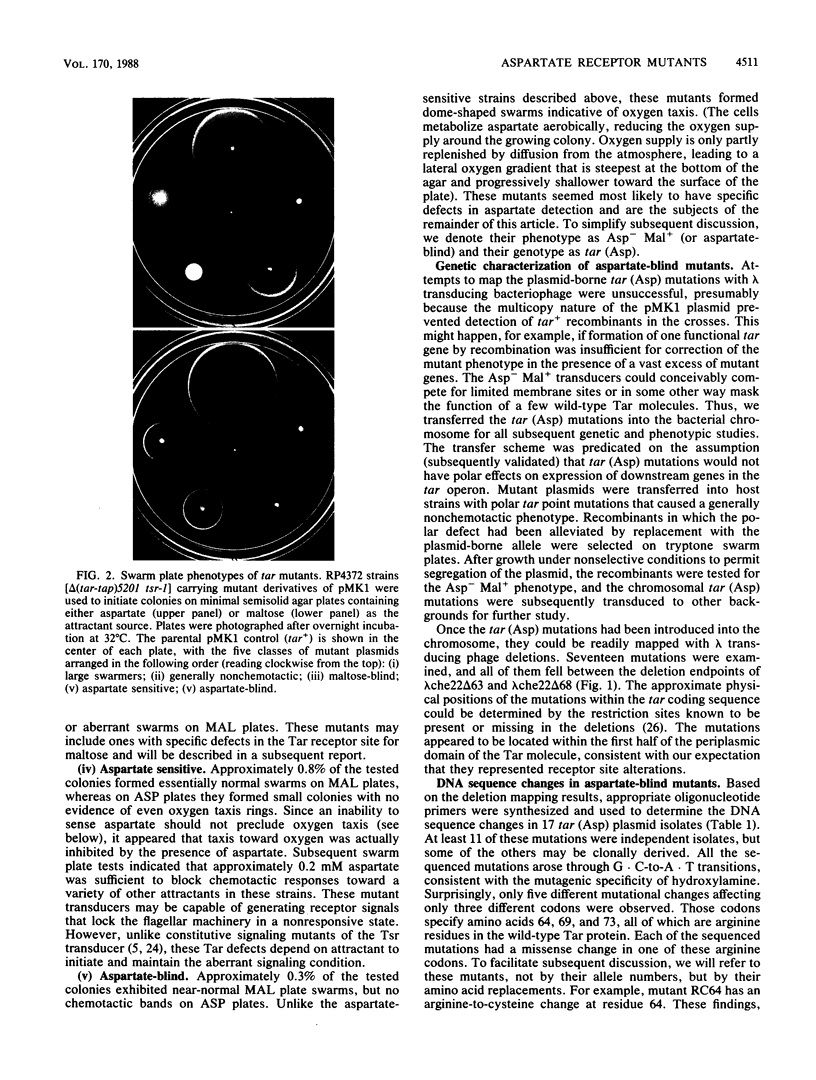
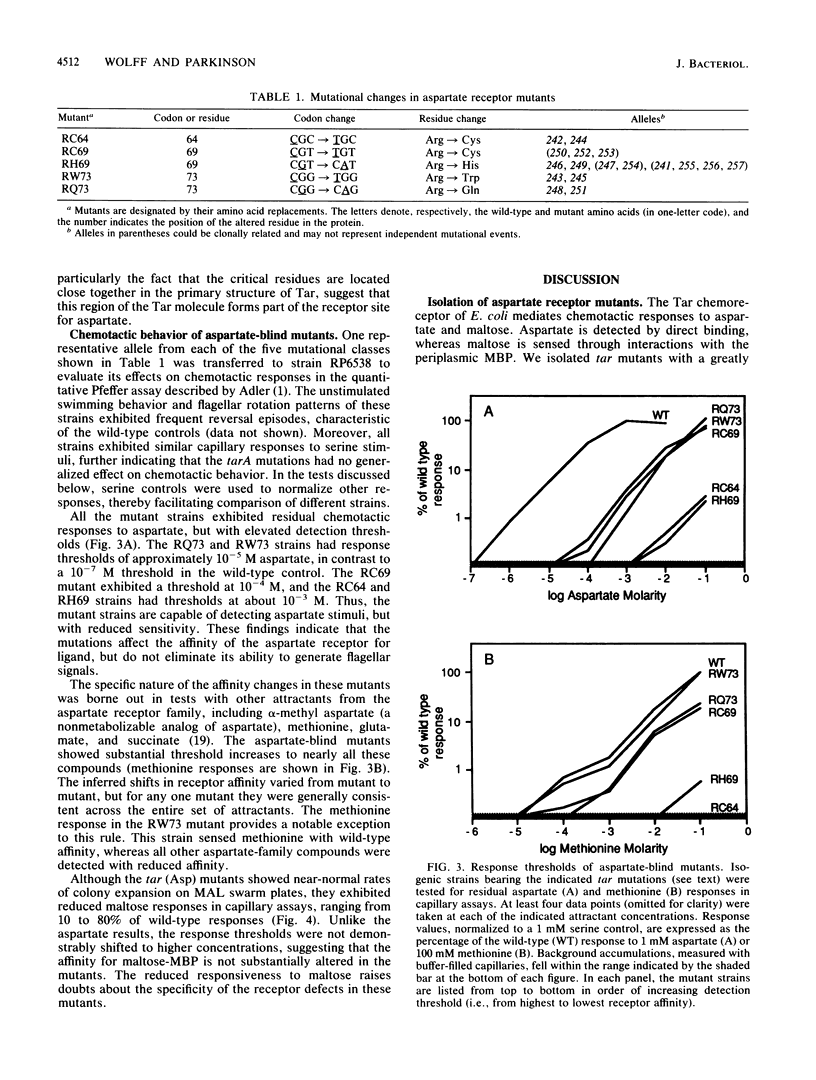
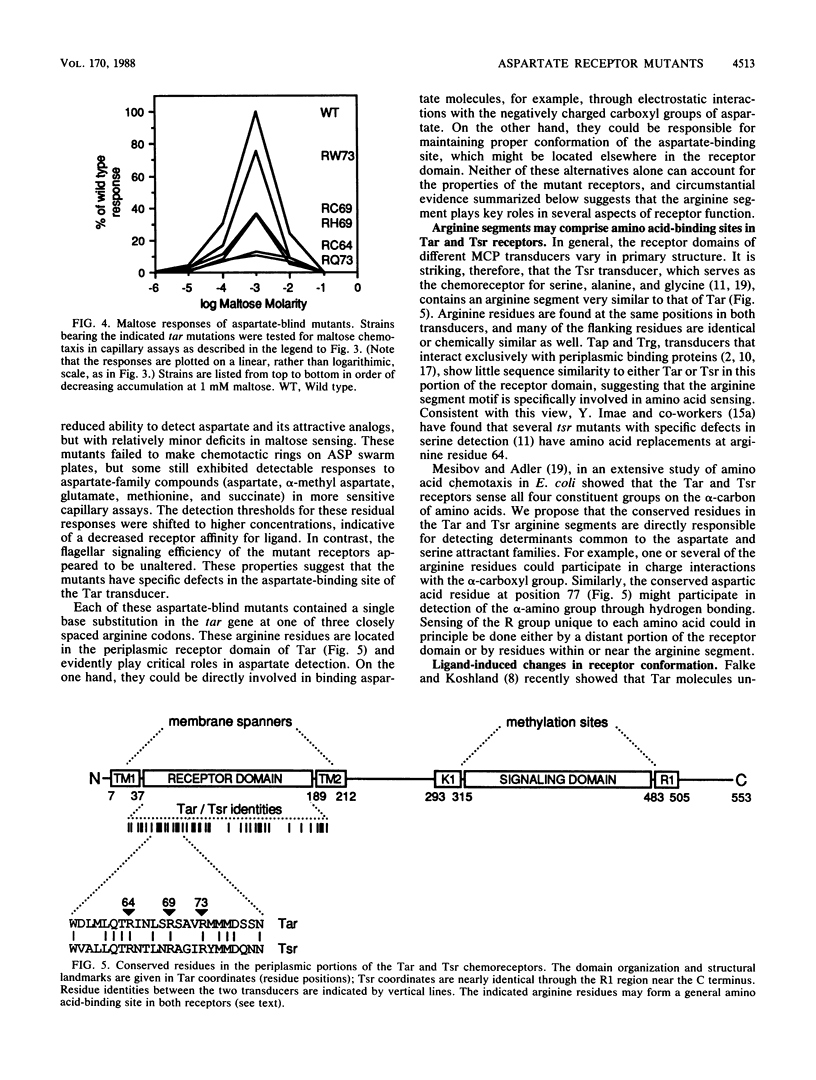
Abstract
The Tar protein of Escherichia coli belongs to a family of methyl-accepting inner membrane proteins that mediate chemotactic responses to a variety of compounds. These transmembrane signalers monitor the chemical environment by means of specific ligand-binding sites arrayed on the periplasmic side of the membrane, and in turn control cytoplasmic signals that modulate the flagellar rotational machinery. The periplasmic receptor domain of Tar senses two quite different chemoeffectors, aspartate and maltose. Aspartate is detected through direct binding to Tar molecules, whereas maltose is detected indirectly when complexed with the periplasmic maltose-binding protein. Saturating levels of either aspartate or maltose do not block behavioral responses to the other compound, indicating that the detection sites for these two attractants are not identical. We initiated structure-function studies of these chemoreceptor sites by isolating tar mutants which eliminate aspartate or maltose taxis, while retaining the ability to respond to the other chemoeffector. Mutants with greatly reduced aspartate taxis are described and characterized in this report. When present in single copy in the chromosome, these tar mutations generally eliminated chemotactic responses to aspartate and structurally related compounds, such as glutamate and methionine. Residual responses to these compounds were shifted to higher concentrations, indicating a reduced affinity of the aspartate-binding site in the mutant receptors. Maltose responses in the mutants ranged from 10 to 80% of normal, but had no detectable threshold shifts, indicating that these receptor alterations may have little effect on maltose detection sensitivity. The mutational changes in 17 mutants were determined by DNA sequence analysis. Each mutant exhibited a single amino acid replacement at residue 64, 69, or 73 in the Tar molecule. The wild-type Tar transducer contains arginines at all three of these positions, implying that electrostatic forces may play an important role in aspartate detection.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973 Jan;74(1):77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Aksamit R. R., Koshland D. E., Jr Identification of the ribose binding protein as the receptor for ribose chemotaxis in Salmonella typhimurium. Biochemistry. 1974 Oct 22;13(22):4473–4478. doi: 10.1021/bi00719a001. [DOI] [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Callahan A. M., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: cheD mutations affect the structure and function of the Tsr transducer. J Bacteriol. 1985 Jan;161(1):96–104. doi: 10.1128/jb.161.1.96-104.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun S. Y., Parkinson J. S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988 Jan 15;239(4837):276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- Débarbouillé M., Shuman H. A., Silhavy T. J., Schwartz M. Dominant constitutive mutations in malT, the positive regulator gene of the maltose regulon in Escherichia coli. J Mol Biol. 1978 Sep 15;124(2):359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- Falke J. J., Koshland D. E., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987 Sep 25;237(4822):1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Adler J. Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose. Nat New Biol. 1971 Mar 24;230(12):101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L. Maltose chemoreceptor of Escherichia coli. J Bacteriol. 1975 Apr;122(1):206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedblom M. L., Adler J. Genetic and biochemical properties of Escherichia coli mutants with defects in serine chemotaxis. J Bacteriol. 1980 Dec;144(3):1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann O., Szmelcman S. Active transport of maltose in Escherichia coli K12. Involvement of a "periplasmic" maltose binding protein. Eur J Biochem. 1974 Aug 15;47(1):139–149. doi: 10.1111/j.1432-1033.1974.tb03677.x. [DOI] [PubMed] [Google Scholar]
- Koiwai O., Hayashi H. Studies on bacterial chemotaxis. IV. Interaction of maltose receptor with a membrane-bound chemosensing component. J Biochem. 1979 Jul;86(1):27–34. [PubMed] [Google Scholar]
- Kossmann M., Wolff C., Manson M. D. Maltose chemoreceptor of Escherichia coli: interaction of maltose-binding protein and the tar signal transducer. J Bacteriol. 1988 Oct;170(10):4516–4521. doi: 10.1128/jb.170.10.4516-4521.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Lee L., Mizuno T., Imae Y. Thermosensing properties of Escherichia coli tsr mutants defective in serine chemoreception. J Bacteriol. 1988 Oct;170(10):4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manson M. D., Blank V., Brade G., Higgins C. F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986 May 15;321(6067):253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Boos W., Bassford P. J., Jr, Rasmussen B. A. Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J Biol Chem. 1985 Aug 15;260(17):9727–9733. [PubMed] [Google Scholar]
- Mesibov R., Adler J. Chemotaxis toward amino acids in Escherichia coli. J Bacteriol. 1972 Oct;112(1):315–326. doi: 10.1128/jb.112.1.315-326.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowbray S. L., Foster D. L., Koshland D. E., Jr Proteolytic fragments identified with domains of the aspartate chemoreceptor. J Biol Chem. 1985 Sep 25;260(21):11711–11718. [PubMed] [Google Scholar]
- Mowbray S. L., Koshland D. E., Jr Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell. 1987 Jul 17;50(2):171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Complementation analysis and deletion mapping of Escherichia coli mutants defective in chemotaxis. J Bacteriol. 1978 Jul;135(1):45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S. Novel mutations affecting a signaling component for chemotaxis of Escherichia coli. J Bacteriol. 1980 Jun;142(3):953–961. doi: 10.1128/jb.142.3.953-961.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richarme G. Interaction of the maltose-binding protein with membrane vesicles of Escherichia coli. J Bacteriol. 1982 Feb;149(2):662–667. doi: 10.1128/jb.149.2.662-667.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Silverman M., Simon M. Chemotaxis in Escherichia coli: methylation of che gene products. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3317–3321. doi: 10.1073/pnas.74.8.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: null phenotypes of the tar and tap genes. J Bacteriol. 1985 Aug;163(2):586–594. doi: 10.1128/jb.163.2.586-594.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983 Aug;155(2):565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T. C., Koshland D. E., Jr Sites of methyl esterification and deamination on the aspartate receptor involved in chemotaxis. J Biol Chem. 1984 Jun 25;259(12):7719–7725. [PubMed] [Google Scholar]
- Wang E. A., Koshland D. E., Jr Receptor structure in the bacterial sensing system. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7157–7161. doi: 10.1073/pnas.77.12.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]