Abstract
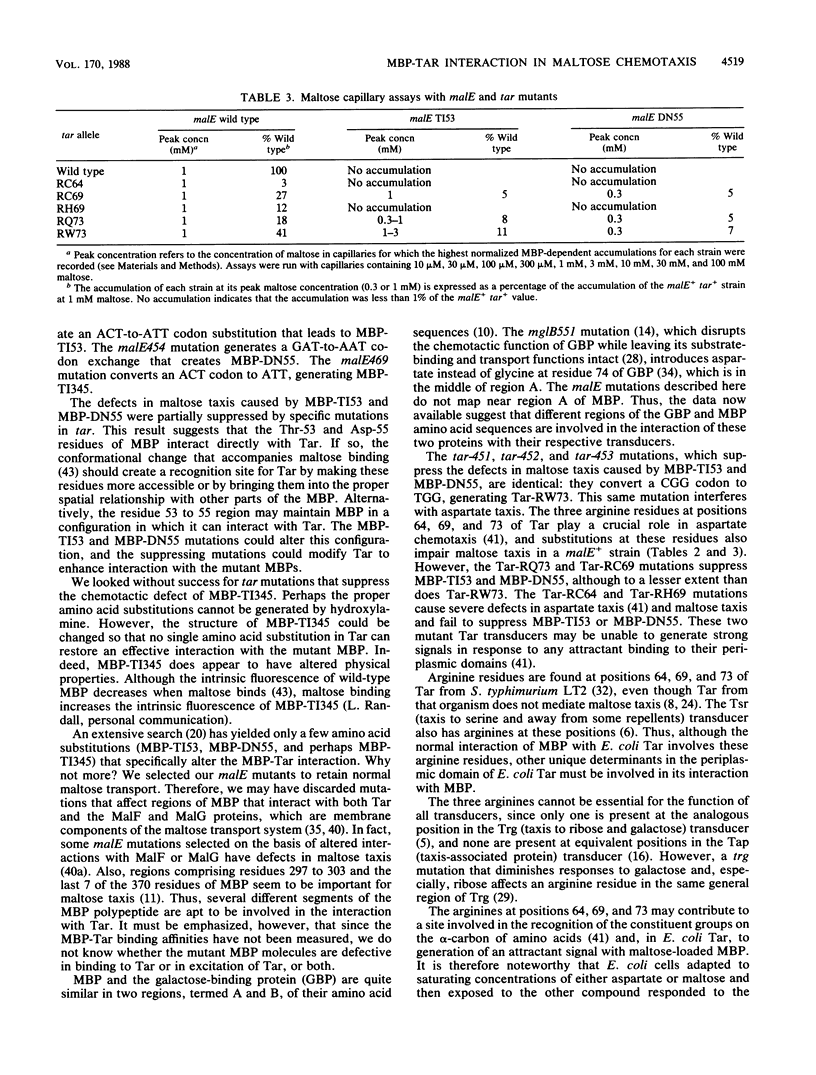
The maltose chemoreceptor in Escherichia coli consists of the periplasmic maltose-binding protein (MBP) and the Tar signal transducer, which is localized in the cytoplasmic membrane. We previously isolated strains containing malE mutations that cause specific defects in the chemotactic function of MBP. Four of these mutations have now been characterized by DNA sequence analysis. Two of them replace threonine at residue 53 of MBP with isoleucine (MBP-TI53), one replaces an aspartate at residue 55 with asparagine (MBP-DN55), and the fourth replaces threonine at residue 345 with isoleucine (MBP-TI345). The chemotactic defects of MBP-TI53 and MBP-DN55, but not of MBP-TI345, are suppressed by mutations in the tar gene. Of the tar mutations, the most effective suppressor (isolated independently three times) replaces Arg-73 of Tar with tryptophan. Two other tar mutations that disrupt the aspartate chemoreceptor function of Tar also suppress the maltose taxis defects associated with MBP-TI53 and MBP-DN55. One of these mutations introduces glutamine at residue 73 of Tar, the other replaces arginine at residue 69 of Tar with cysteine. These results suggest that regions of MBP that include residues 53 to 55 and residue 345 are important for the interaction with Tar. In turn, arginines at residues 69 and 73 of Tar must be involved in the recognition of maltose-bound MBP and/or in the production of the attractant signal generated by Tar in response to maltose-bound MBP.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. A method for measuring chemotaxis and use of the method to determine optimum conditions for chemotaxis by Escherichia coli. J Gen Microbiol. 1973 Jan;74(1):77–91. doi: 10.1099/00221287-74-1-77. [DOI] [PubMed] [Google Scholar]
- Adler J., Hazelbauer G. L., Dahl M. M. Chemotaxis toward sugars in Escherichia coli. J Bacteriol. 1973 Sep;115(3):824–847. doi: 10.1128/jb.115.3.824-847.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedouelle H., Bassford P. J., Jr, Fowler A. V., Zabin I., Beckwith J., Hofnung M. Mutations which alter the function of the signal sequence of the maltose binding protein of Escherichia coli. Nature. 1980 May 8;285(5760):78–81. doi: 10.1038/285078a0. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bollinger J., Park C., Harayama S., Hazelbauer G. L. Structure of the Trg protein: Homologies with and differences from other sensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3287–3291. doi: 10.1073/pnas.81.11.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Dahl M. K., Manson M. D. Interspecific reconstitution of maltose transport and chemotaxis in Escherichia coli with maltose-binding protein from various enteric bacteria. J Bacteriol. 1985 Dec;164(3):1057–1063. doi: 10.1128/jb.164.3.1057-1063.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duplay P., Bedouelle H., Fowler A., Zabin I., Saurin W., Hofnung M. Sequences of the malE gene and of its product, the maltose-binding protein of Escherichia coli K12. J Biol Chem. 1984 Aug 25;259(16):10606–10613. [PubMed] [Google Scholar]
- Duplay P., Szmelcman S. Silent and functional changes in the periplasmic maltose-binding protein of Escherichia coli K12. II. Chemotaxis towards maltose. J Mol Biol. 1987 Apr 20;194(4):675–678. doi: 10.1016/0022-2836(87)90244-0. [DOI] [PubMed] [Google Scholar]
- Débarbouillé M., Shuman H. A., Silhavy T. J., Schwartz M. Dominant constitutive mutations in malT, the positive regulator gene of the maltose regulon in Escherichia coli. J Mol Biol. 1978 Sep 15;124(2):359–371. doi: 10.1016/0022-2836(78)90304-2. [DOI] [PubMed] [Google Scholar]
- Falke J. J., Koshland D. E., Jr Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science. 1987 Sep 25;237(4822):1596–1600. doi: 10.1126/science.2820061. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L., Adler J. Role of the galactose binding protein in chemotaxis of Escherichia coli toward galactose. Nat New Biol. 1971 Mar 24;230(12):101–104. doi: 10.1038/newbio230101a0. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L. Maltose chemoreceptor of Escherichia coli. J Bacteriol. 1975 Apr;122(1):206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellermann O., Szmelcman S. Active transport of maltose in Escherichia coli K12. Involvement of a "periplasmic" maltose binding protein. Eur J Biochem. 1974 Aug 15;47(1):139–149. doi: 10.1111/j.1432-1033.1974.tb03677.x. [DOI] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Manson M. D., Boos W., Bassford P. J., Jr, Rasmussen B. A. Dependence of maltose transport and chemotaxis on the amount of maltose-binding protein. J Biol Chem. 1985 Aug 15;260(17):9727–9733. [PubMed] [Google Scholar]
- Manson M. D., Kossmann M. Mutations in tar suppress defects in maltose chemotaxis caused by specific malE mutations. J Bacteriol. 1986 Jan;165(1):34–40. doi: 10.1128/jb.165.1.34-40.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Mizuno T., Mutoh N., Panasenko S. M., Imae Y. Acquisition of maltose chemotaxis in Salmonella typhimurium by the introduction of the Escherichia coli chemosensory transducer gene. J Bacteriol. 1986 Mar;165(3):890–895. doi: 10.1128/jb.165.3.890-895.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowbray S. L., Foster D. L., Koshland D. E., Jr Proteolytic fragments identified with domains of the aspartate chemoreceptor. J Biol Chem. 1985 Sep 25;260(21):11711–11718. [PubMed] [Google Scholar]
- Mowbray S. L., Koshland D. E., Jr Additive and independent responses in a single receptor: aspartate and maltose stimuli on the tar protein. Cell. 1987 Jul 17;50(2):171–180. doi: 10.1016/0092-8674(87)90213-3. [DOI] [PubMed] [Google Scholar]
- Mutoh N., Oosawa K., Simon M. I. Characterization of Escherichia coli chemotaxis receptor mutants with null phenotypes. J Bacteriol. 1986 Sep;167(3):992–998. doi: 10.1128/jb.167.3.992-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordal G. W., Adler J. Properties of mutants in galactose taxis and transport. J Bacteriol. 1974 Feb;117(2):517–526. doi: 10.1128/jb.117.2.517-526.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridgway H. G., Silverman M., Simon M. I. Localization of proteins controlling motility and chemotaxis in Escherichia coli. J Bacteriol. 1977 Nov;132(2):657–665. doi: 10.1128/jb.132.2.657-665.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle A., Vreemann J., Blank V., Nold A., Boos W., Manson M. D. Sequence of the mglB gene from Escherichia coli K12: comparison of wild-type and mutant galactose chemoreceptors. Mol Gen Genet. 1987 Jun;208(1-2):247–253. doi: 10.1007/BF00330450. [DOI] [PubMed] [Google Scholar]
- Shuman H. A. Active transport of maltose in Escherichia coli K12. Role of the periplasmic maltose-binding protein and evidence for a substrate recognition site in the cytoplasmic membrane. J Biol Chem. 1982 May 25;257(10):5455–5461. [PubMed] [Google Scholar]
- Silhavy T. J., Brickman E., Bassford P. J., Jr, Casadaban M. J., Shuman H. A., Schwartz V., Guarente L., Schwartz M., Beckwith J. R. Structure of the malB region in Escherichia coli K12. II. Genetic map of the malE,F,G operon. Mol Gen Genet. 1979 Jul 24;174(3):249–259. doi: 10.1007/BF00267797. [DOI] [PubMed] [Google Scholar]
- Slocum M. K., Parkinson J. S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983 Aug;155(2):565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treptow N. A., Shuman H. A. Allele-specific malE mutations that restore interactions between maltose-binding protein and the inner-membrane components of the maltose transport system. J Mol Biol. 1988 Aug 20;202(4):809–822. doi: 10.1016/0022-2836(88)90560-8. [DOI] [PubMed] [Google Scholar]
- Treptow N. A., Shuman H. A. Genetic evidence for substrate and periplasmic-binding-protein recognition by the MalF and MalG proteins, cytoplasmic membrane components of the Escherichia coli maltose transport system. J Bacteriol. 1985 Aug;163(2):654–660. doi: 10.1128/jb.163.2.654-660.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff C., Parkinson J. S. Aspartate taxis mutants of the Escherichia coli tar chemoreceptor. J Bacteriol. 1988 Oct;170(10):4509–4515. doi: 10.1128/jb.170.10.4509-4515.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zukin R. S. Evidence for a conformational change in the Escherichia coli maltose receptor by excited-state fluorescence lifetime data. Biochemistry. 1979 May 29;18(11):2139–2145. doi: 10.1021/bi00578a001. [DOI] [PubMed] [Google Scholar]


