Abstract
To identify sites of action of volatile anesthetics, we are studying genes in a functional pathway that controls sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans. The unc-1 gene occupies a central position in this pathway. Different alleles of unc-1 have unique effects on sensitivity to the different volatile anesthetics. UNC-1 shows extensive homology to human stomatin, an integral membrane protein thought to regulate an associated ion channel. We postulate that UNC-1 has a direct effect on anesthetic sensitivity in C. elegans and may represent a molecular target for volatile anesthetics.
Volatile anesthetics have revolutionized the practice of medicine, yet no one understands how they produce their desired effects (1–3). Whatever the nature of specific anesthetic targets, volatile anesthetics appear to disrupt fundamental aspects of neuronal function, ones that appear to be highly conserved across many disparate phyla (4). Such basic functions are ideally suited to genetic analysis in a simple animal model. To identify sites of anesthetic action, we have undertaken a study of genes that control sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans (5).
Since the turn of the century it has been known that, in all animals studied, the potency of a volatile anesthetic is primarily a function of its oil/gas partition coefficient (6, 7). This relationship, termed the Meyer–Overton rule, states that the product of the oil–gas partition coefficient (in olive oil or octanol) and the EC50 (the effective concentration at which 50% of animals are anesthetized) is approximately a constant for all the volatile anesthetics. We have shown (5) that C. elegans adheres to the Meyer–Overton rule and responds to volatile anesthetics in a manner similar to mammals. Historically, the Meyer–Overton rule has been interpreted to mean that the anesthetic site of action resembles olive oil or octanol (4) and that there is one type of site of action for all volatile anesthetics in all species (termed the unitary hypothesis) (4, 8).
The unitary hypothesis, in its simplest form, predicts that mutations at anesthetic sites of action should affect sensitivities to all volatile anesthetics in a similar fashion. However, genetic studies in several model systems have identified mutations that affect sensitivities to different volatile anesthetics in different ways. We have found mutations in C. elegans that only alter sensitivity to specific volatile anesthetics, whereas other mutations confer hypersensitivity to all of them (5, 9, 10). Mutations in the unc-1 gene represent one example of a genetic change affecting some anesthetic sensitivities and not others. Independent studies in Drosophila and in mice also indicate that genetic changes can alter sensitivity to some volatile anesthetics and not others (11–14). Kendig et al. (15) have found differences between various volatile anesthetics in neurophysiologic studies of the hippocampus of rats. These data as a whole indicate that the unitary hypothesis is probably an oversimplification and that the mechanism of action of volatile anesthetics may involve several molecular sites.
Using immobility as an endpoint, eight genes have been identified that affect sensitivity to volatile anesthetics in C. elegans (5, 10, 16). The interactions between mutations in these genes suggest that they lie in a functional pathway controlling anesthetic sensitivity. The gene unc-1 occupies a central position in this pathway. Loss of function mutations in unc-1 suppress the altered sensitivities of upstream mutations (unc-79) and confer their own changes in sensitivities to volatile anesthetics (5). When not exposed to anesthetics, unc-1 loss of function alleles do not have the normal sinusoidal motion seen in the wild-type nematode; instead, they exhibit a motion described as kinked. The characterization of unc-1 serves as a key in understanding the action of volatile anesthetics.
METHODS
Strains and General Methods.
Basic genetic techniques and those for culturing C. elegans along with genetic and physical mapping were as described by Brenner (17) and Williams (18). Molecular biology techniques were done by standard procedures except as noted. Certain nematode strains were obtained from the Caenorhabditis Genetics Center in Minneapolis, MN. Other strains used in these experiments were kindly provided to us by the following investigators. The unc-1 alleles n494 and n774 (Carl Johnson, Nemapharm, Boston, MA); the unc-1 alleles hs1, hs2, hs3, hs4, and hs5 (Ralph Hecht, Univ. of Houston, Houston, TX); unc-1(e1598n1201)dpy3 (Anne Villeneuve, Stanford Univ. Medical School, San Francisco, CA); and RW7000 (Bertold Schrank, Washington Univ., St. Louis, MO). fc53 was obtained spontaneously from an UncnonDpy animal identified in an F1 noncomplementation screen when unc-1(e1598n1201)dpy3 animals were crossed with the mutator strain RW7097. Procedures for exposing nematodes to anesthetics, scoring their responses, and measuring the anesthetic concentrations have been described in detail (5, 9).
Cosmids and Mutant Rescue.
Cosmids from the region of the unc-1 gene were provided by Alan Coulson (The Sanger Centre, Cambridge, U.K.). Cosmid DNA was prepared with the Plasmid Maxiprep kit (Qiagen, Chatsworth, CA). Mutant rescue was done as described by Fire (19) and by Mello et al. (20). Briefly, this technique involves injecting wild-type DNA into the gonad of mutant animals. The injected DNA is taken up by the developing oocytes and can form free linear arrays. If the injected DNA contains a wild-type copy of the mutated gene, then some of the offspring of the mutant parent will be wild type; i.e., they will be “rescued.” Loss of function mutants of unc-1 have an abnormal “kinked” motion. Rescued progeny of unc-1(0) parents will move normally. In general, the test DNA was injected at 10 μg/ml; a marker for successful microinjection, rol-6 DNA [pRF4, a plasmid containing the dominant rol-6(su1006) mutation] was coinjected at 100 μg/ml. F1 Rollers were picked to establish stable lines. Stable lines were scored for loss of the kinked Unc-1 phenotype described above and also for their anesthetic response.
cDNA.
On the basis of Genefinder predictions, oligonucleotides were synthesized (Genosys, The Woodlands, TX) from putative exons of the K03E6.3 and K03E6.5 genes with maximum homology to neurocalcin and stomatin, respectively. These were used to screen mixed stage C. elegans cDNA libraries (provided by R. Barstead, Oklahoma Medical Research Foundation, Oklahoma City, OK, and P. Okkema, Univ. of Illinois at Chicago, Chicago, IL). Original libraries were amplified once and then subjected to 30 cycles of PCR under standard conditions, by using Taq polymerase (GIBCO/BRL.). Fragments thus amplified were sequenced to verify that they contained sequences from K03E6.3 and K03E6.5, respectively, and to know the true intron–exon boundaries for each gene.
Rapid Amplification of cDNA Ends.
Two micrograms of DNase-treated total RNA was reverse-transcribed with 25 pmol of either a gene-specific or an oligo(dT)/adapter primer, essentially by the method of Frohman et al. (21). Moloney murine leukemia virus reverse transcriptase (GIBCO/BRL) and the buffer supplied with it were used in the reaction. DTT (1 mM), all four dNTPs (each at 1 mM), and RNAsin (20 units) (Promega Biotech) were added separately. Two microliters of the products from this reverse transcriptase reaction was then subjected to 30 cycles of amplification by using nested gene-specific primers (annealing temperature; 58°C) and the Expand High Fidelity PCR system (Boehringer Mannheim Biochemicals). Buffers and reaction conditions were as supplied and recommended by the manufacturer.
The 5′ ends of the K03E6.5 and K03E6.3 transcripts were determined by reverse transcription-coupled PCR analysis using the two C. elegans splice leader sequence primers SL1 (5′-GGTTTAATTACCCAAGTTTGAG-3′) and SL2 (5′-GGTTTTAACCCAGTTACTCAAG-3′) (22). In each case, no PCR products were detected with SL2 under the conditions used but single bands were isolated when SL1 was used, indicating that both messages were SL1 trans-spliced. The PCR products were sequenced to ascertain that they truly represented transcripts of the K03E6.5 and K03E6.3 genes.
Sequencing.
Reverse transcription-coupled PCR was used to generate cDNA fragments corresponding to the mRNA from K03E6.3 and K03E6.5. The reverse transcription reaction was primed with random hexamers. DNase-treated RNA from N2 and each of the five unc-1 alleles (n494, n774, e114, e580, and hs1) was used for these experiments. Resulting fragments were amplified by using primers specific to each gene and Expand High Fidelity PCR system as described above. The K03E6.5 gene was also sequenced from the DNA of the following unc-1 alleles: e719, fc53, e1598n1202, and hs2–hs5. Sequencing was done by the Genetics Sequencing Facilities at Case Western Reserve University and the Cleveland Clinic Foundation. The sequences of the primers used for sequencing are available on request. The frameshift mutations in fc53 and e719 were confirmed by resequencing fragments generated by reverse transcription-coupled PCR. Any mutations identified in the other unc-1 alleles were confirmed by reamplifying that segment at least one more time and all regions were sequenced in both directions.
RESULTS
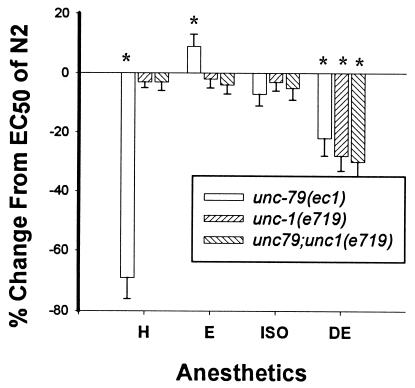
Loss of function mutations in the unc-1 gene restore to normal the hypersensitivity of unc-79 mutants to all the more lipid soluble anesthetics such as halothane (Fig. 1) (5). By themselves, unc-1 loss of function mutants cause a 30% increase in sensitivity specifically to diethyl ether. The dominant unc-1 allele n494 (25) showed an increased sensitivity to halothane, isoflurane, enflurane, and ether. The double mutant unc-79(ec1);n494 is a poor mover and has an increased sensitivity to halothane relative to either mutation alone, consistent with an interaction between these gene products. The double mutant did not show an increased sensitivity to enflurane or isoflurane compared with n494 alone. These results indicate that the UNC-1 protein is affected at low concentrations of halothane and that the interaction of UNC-1 and UNC-79 are important in determining the sensitivity of C. elegans to volatile anesthetics.
Figure 1.
Histogram showing changes in EC50 values of unc-79, unc-1, and unc-79;unc-1 from those of N2 (baseline) in four anesthetics. H, halothane; E, enflurane; ISO, isoflurane; DE, diethyl ether. Note the ability of unc-1 to suppress the increased sensitivities of unc-79 to halothane. unc-1 also suppresses the decreased sensitivities of unc-79 to enflurane. unc-1 by itself has an increased sensitivity to diethyl ether and does not suppress the increased sensitivity of unc-79 to ether. The null allele of unc-1 used in these studies is e719, which behaves like e580 in other comparative tests of phenotype and anesthetic responses. EC50 values, standard errors, and comparisons among strains were calculated as described by Waud (23). For each anesthetic, the EC50 values of all strains were compared by using an analysis of variance (24). Significance was defined as P < 0.01. Those EC50 values significantly different than the EC50 value of N2 are noted with an asterisk (∗). The EC50 values for halothane in the experiments were 3.2% for N2, 0.7% for unc-79, 3.5% for unc-1, and 3.3% for unc-79;unc-1. These values were used as a guide for the concentrations used to test for rescue of unc-1 described in Results.
We identified the unc-1 gene by positional cloning and mutant rescue (19, 20). The cosmid K03E6 rescued the kinked phenotype of e580(0) as well as the dominant coiled phenotype of n494. The smallest fragment of K03E6 capable of rescuing unc-1 is a 5.6-kb HindIII fragment spanning the interval from 0.7 kb to 6.3 kb of the cosmid. This fragment contains portions of two predicted genes transcribed in opposite directions (Fig. 2). The rescuing fragment contains five exons of the predicted gene K03E6.5, which is homologous to stomatin, an integral membrane phosphoprotein (26). It also contains four exons of the gene K03E6.3, which encodes a protein homologous to neurocalcin, a neuron-specific calcium binding protein (27). Smaller fragments, in which the two partial genes were separated, failed to rescue the Unc-1 phenotype.
Figure 2.
Restriction map of the K03E6 cosmid (0–14 kb) showing the positions of the K03E6.5 (stomatin-like; solid boxes) and K03E6.3 (neurocalcin-like; open boxes) coding regions. Arrows indicate the direction of transcription of each gene. The thick black line shows the position of the rescuing 5.6-kb HindIII fragment. The location of the deletions in fc53 [2.3 kb (0.9–2.2 kb) in K03E6.3 and 13 bp in exon 4 of K03E6.5] are shown by the discontinuous line above the cosmid. The first two exons of K03E6.3, which lie outside the K03E6 cosmid, are shown extending beyond the thin black line representing the K03E6 cosmid.
cDNAs corresponding to each of these genes were obtained by screening mixed-stage C. elegans cDNA libraries and were used to probe genomic DNA from the different unc-1 alleles. The fc53 allele (a transposon induced null allele of unc-1) was found to be associated with a 2.3-kb deletion in the K03E6.3 (neurocalcin-like) gene. Sequencing confirmed the location of the deletion breakpoints (Fig. 2). Northern blots showed complete absence of the K03E6.3 message in fc53. Additionally, the K03E6.5 (stomatin-like) message from fc53 showed a 90% decrease in transcription of that gene as well. To identify the true extent of both genes, we performed rapid amplification of cDNA ends and PCR by using primers from predicted exons with strong homology to stomatin and neurocalcin, respectively. Both messages were found to be trans-spliced to an SL1 leader (22). We found that K03E6.5 contains only six of the eight exons predicted by Genefinder. Therefore, the smallest rescuing fragment omitted only the first exon of K03E6.5, corresponding to the N-terminal 18 amino acids of the stomatin-like protein. These are the least conserved across species and, therefore, presumably less important for function. In addition, rapid amplification of cDNA ends and PCR also showed that more than half of the coding region of the neurocalcin-like gene extended beyond K03E6 in a region not sequenced previously by the sequencing consortium. The structure of the neurocalcin-like gene will be reported elsewhere. Thus, these results made it unlikely that unc-1 encoded the neurocalcin homologue. However, because the deletion found in fc53 affected transcription of both genes and the rescuing fragments also contained parts of both genes, we were unable to unequivocally differentiate between the two homologues as the unc-1 gene. Therefore, we sequenced both genes in a representative of each class of unc-1 alleles (e580, e114, n774, and n494) and the temperature-sensitive allele hs1 (28).
The sequences of the neurocalcin-like gene K03E6.3 in the above unc-1 alleles (e580, e114, n774, n494, and hs1) did not differ from the sequence in N2. Conversely, all five alleles show significant changes in the coding sequence of the stomatin-like gene K03E6.5 (Fig. 3). We also sequenced the stomatin-like gene in seven other unc-1 alleles. These included the canonical null allele e719, the remaining four temperature-sensitive alleles hs2-hs5, the transposon-induced allele fc53, and the unc-1(null) parent of fc53 e1598n120 (see Methods). Each of these seven alleles also shows significant changes in K03E6.5. The sequence data from N2 indicated that K03E6.5 spans six exons and contains an ORF of 855 bp, encoding a protein of 285 amino acids with a predicted molecular mass of about 31 kDa. By using these data to predict where the promoter and polyadenylation sites were, we were able to rescue both the kinked and altered anesthetic sensitivities characteristics of the Unc-1 phenotype (using e580) with a genomic fragment containing only sequence from K03E6.5 subcloned into pBluescript. All rolling nonkinked rescued animals were insensitive to 6.5% diethyl ether (like N2). In addition, when the double mutant unc-79;unc-1(e580) was injected with this plasmid, the F1 offspring were not kinked. Stable lines were obtained from these rescued worms and exposed to different doses of halothane. All (25 of 25) rolling animals were immobilized by 0.9% halothane. (The control unc-79 single mutant was completely immobilized at 0.9% halothane. The EC50s for N2, unc-79, unc-1, and unc-79;unc-1 are given in Fig. 1.) We therefore conclude that unc-1 is the stomatin-like gene K03E6.5 and mediates both changes in anesthetic sensitivity and kinked motion.
Figure 3.
Multiple alignment of the deduced amino acid sequence of UNC-1 with that of the two mammalian stomatins (15). M, mouse; H, human; *, identity; dot, similarity. Most of the unc-1 mutations (shown above the boldfaced type) lie in residues that are identical or similar between the proteins (★, stop). The two ⇓ marks indicate the region that is replaced with the following residues RTCGSEGHPSPPTTHTCHGRRSRGRA in e719. The predicted membrane-spanning domain for each protein is shaded (22). A second membrane spanning domain is also predicted for the UNC-1 protein. Cysteine residues within the hydrophobic stretch that may potentially be palmitoylated to provide membrane-anchoring function are underlined. The 1.2-kb K03E6.5 transcript includes a 3′ untranslated region of 343 bp in addition to an ORF of 855 bp (29).
DISCUSSION
The unc-1 gene has unique effects on anesthetic sensitivity; kinked, loss of function mutations decrease sensitivity of unc-79 mutants to halothane and, by themselves, increase sensitivity to ether. Dominant unc-1 mutations increase sensitivity to all volatile anesthetics. These results indicate that the Unc-1 protein is important in the response of C. elegans to volatile anesthetics. The identification of Unc-1 as a homologue to a human membrane protein affecting an associated ion channel represents a major clue as to how these anesthetics function.
The UNC-1 protein has two hydrophobic regions near its N terminus; its N and C termini are both hydrophilic and are predicted be intracellular. As shown in Fig. 3, the homology between UNC-1 and the two mammalian stomatin proteins is extensive (52% identity and 87% similarity overall; 58% identity and 91% similarity over the regulatory cytoplasmic region) (26). Given the predicted similarities between these proteins, we believe that their functions are probably similar. In humans, absence of stomatin from erythrocyte membranes results in anemia, overhydrated hereditary stomatocytosis (OHSt) (26). The red blood cell membranes of OHSt patients show increased permeabilities to Na+ and K+ ions, resulting in swelling and lysis of the red blood cells due to a reversal of normal intracellular Na+/K+ ratios. Stomatin is therefore believed to regulate an associated ion channel, probably a sodium channel, in a “ball and chain” manner (29, 30). Likewise, if UNC-1 regulates ion flow, then it may be responsible for maintaining the ionic gradient in the cells in which it is expressed. Preliminary results with green fluorescent protein expression (31) under the control of the unc-1 promoter shows intense expression of the gene broadly within the nervous system and little expression outside of nerve cells (S.R., unpublished results).
There are two other stomatin-like proteins, MEC-2 and UNC-24, in C. elegans (32, 33). Both proteins are only partially homologous to stomatin and UNC-1. Mutations in the mec-2 gene do not alter sensitivity to anesthetics, but mutations in unc-24 do partially suppress the anesthetic sensitivity of unc-79 (5). Between these two proteins, however, UNC-1 is more homologous to MEC-2 than to UNC-24. mec-2 alleles are deficient in the control of mechanosensation in C. elegans. In C. elegans, MEC-2 regulates an ion channel necessary for mechanosensation (34). This ion channel belongs to the class of amiloride-sensitive epithelial sodium channels (35, 36).
All of the unc-1 alleles that we have characterized are altered in residues that are well conserved between the stomatins of different species (Fig. 3) (29, 30). Thus far, the sequence data has shown the following structure–function relationships. All of the temperature-sensitive alleles are the result of changes close to the C-terminal end, in the region predicted to form the “ball” that gates the associated ion channel by acting as a plug (Fig. 4). The remaining mutations that retain some function result from changes in the proposed large cytoplasmic region, presumably in the “chain.” Only n774 results from changes close to the proposed membrane spanning regions. As noted in the Introduction, loss of function mutations in unc-1 have a kinked motion. Because n774 is kinked as a homozygote, as is n774/e580(0) (25), it is likely that this allele has little or no normal function. The severe truncations of the cytoplasmic region of the unc-1 protein seen in e719 and fc53 are consistent with the null phenotype of these alleles. Lastly, the N terminus is not necessary for function because constructs lacking this region are able to fully rescue the Unc-1 phenotype. There are more than 30 known alleles of unc-1, and additional sequencing of some of these alleles may reveal changes in the membrane-spanning regions of UNC-1. Such alleles may reveal additional phenotypes when exposed to volatile anesthetics.
Figure 4.
Schematic representation of the way in which UNC-1 may affect membrane conductance and anesthetic sensitivity. Volatile anesthetics such as halothane (H) may bind directly to UNC-1 or to a protein that associates with UNC-1 such as the ion channel that UNC-1 regulates. Altered interactions with any of these proteins in the different unc-1 mutants would then lead to abnormal Na+/K+ fluxes in the affected cells thereby altering their normal motion and affecting their responses to volatile anesthetics. The regions changed by mutations in the 12 sequenced alleles are designated by an asterisk. Note that each asterisk may represent more than one allele; for instance, five different alleles have changes in the proposed “ball,” which is thought to plug an associated channel.
We are interested in how unc-1 mutations affect anesthetic sensitivity. In humans, stomatin is known to bind to lipophilic molecules such as quinolines (37). Thus, UNC-1 represents a potential direct target for lipophilic molecules such as volatile anesthetics. It is possible that normal neuronal function is disrupted by either inactivation or potentiation of UNC-1 by volatile anesthetics. If anesthetics act via inhibition of UNC-1 function, then unc-1 hypomorphs should be sensitive to all volatile anesthetics, and unc-1 nulls should be immobilized in air. Neither is true. If anesthetics potentiate UNC-1, then the gain of function alleles might mimic the effects of volatile anesthetics. An animal carrying the dominant allele n494 is a sluggish mover in air and has an increased sensitivity to all volatile anesthetics, and the allele has additive effects with unc-79 with respect to movement in air and sensitivity to halothane. Because the cosmid K03E6 rescued the coiled phenotype of n494, we think that n494 is a dominant negative allele. It seems most likely that this allele produces a protein that has an opposite effect on UNC-1 function than a loss of function allele; if loss of UNC-1 increases Na+/K+ flux, then n494 decreases it. Rescue of n494 presumably occurs because multiple copies of the wild-type gene are supplied by the cosmid during microinjection. These would compete with the n494 protein product to gate the appropriate channel. The simplest interpretation of the above data is that halothane potentiates the function of UNC-1.
Because it is likely that UNC-1 serves an inhibitory function (i.e., decreasing ion flux), potentiation of UNC-1 would be functionally similar to the effect postulated for volatile anesthetics on the type A γ-aminobutyric acid receptor (1, 3). UNC-1 may determine a major inhibitory effect involved in the graded transmembrane potentials seen in C. elegans by inhibiting ion flow in neurons. In this model, UNC-1 would exert its inhibitory affect on propagation of membrane depolarization whereas the γ-aminobutyric acid receptor exerts its inhibitory effect on synaptic transmission. The possibility that volatile anesthetics may potentiate two very different inhibitory mechanisms may offer a fascinating clue into their function.
It is possible that UNC-1 interacts with other proteins that are the direct targets of volatile anesthetics. Stomatin’s proposed mode of action is that of gating ion flux through interaction with ion channels. Therefore, UNC-1 may play an indirect role in the control of anesthetic sensitivity and the putative ion channel(s) are also candidate(s) for a direct interaction with volatile anesthetics. The other suppressors of unc-79, unc-7, and unc-9, have been identified and are postulated to code for invertebrate gap junction proteins (38, 39). Mutations in these proteins may disrupt UNC-1 function or may independently affect the same physiologic process as does UNC-1. Because no alleles of unc-7 or unc-9 have been found that increase their sensitivity to volatile anesthetics [like unc-1(n494)], it is unclear whether they are integral to anesthetic action.
It is not immediately clear why the effects of unc-1 mutations (alone or as suppressors) are different for different anesthetics. However, because multiple genetic studies have shown that mutations usually affect sensitivity to some volatile anesthetics differently than others (9–13), it is possible that different anesthetics cause “anesthesia” by different mechanisms. Whatever the role of unc-1 in the control of anesthetic sensitivity, the identification of the genes governing anesthetic response represents a crucial first step in understanding how volatile anesthetics work. The fact that a membrane-bound modulator of ion channels is implicated in the control of sensitivity to volatile anesthetics is clearly in support of the protein binding model put forth by Franks and Lieb (1, 40), as well as the observations by others that ion channel currents are affected by volatile anesthetics. We are currently searching for other members of the pathway controlling sensitivity to volatile anesthetics in C. elegans that are genetically downstream of unc-1 to identify possible ion channels and other modulators of anesthetic function. Although a full understanding of this pathway involves identification of several genes, it is unlikely that the control of anesthetic sensitivity is less complicated in mammals than in the nematode.
Acknowledgments
We thank Jim Rand, Carl Johnson, Ann Villeneuve, Tom Barnes, and Ralph Hecht for sharing unpublished data and unc-1 alleles; Susan Thomas and Ted Spangler for their excellent assistance with laboratory procedures; and Peter Harte and Helen Salz for their critical reading of the manuscript. We also thank Helmut Cascorbi for his continuing advice and support and Gordon Stewart and Patrick Gallagher for their helpful discussions. P.G.M. and M.M.S. were supported in part by National Institutes of Health Grant GM45402.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Franks N P, Lieb W R. Nature (London) 1994;367:607–614. doi: 10.1038/367607a0. [DOI] [PubMed] [Google Scholar]
- 2.Gruner S M, Shyamsunder E. Ann NY Acad Sci. 1991;625:685–697. doi: 10.1111/j.1749-6632.1991.tb33902.x. [DOI] [PubMed] [Google Scholar]
- 3.Mihic S J, Ye Q, Wick M J, Koltchine W, Krasowski M D, et al. Nature (London) 1997;389:385–389. doi: 10.1038/38738. [DOI] [PubMed] [Google Scholar]
- 4.Koblin D D. In: Anesthesia. 4th Ed. Miller R D, editor. New York: Churchill Livingstone; 1990. pp. 67–99. [Google Scholar]
- 5.Morgan P G, Sedensky M M, Meneely P M. Proc Natl Acad Sci USA. 1990;87:2965–2969. doi: 10.1073/pnas.87.8.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyer H H. Arch Exp Pathol Pharmakol. 1899;42:109–119. [Google Scholar]
- 7.Overton, E. (1901) Jena Verlag Von Gustav Fisher.
- 8.Tanifuji Y, Eger E I, II, Terrell R C. Anesth Analg. 1977;56:387–391. doi: 10.1213/00000539-197705000-00014. [DOI] [PubMed] [Google Scholar]
- 9.Morgan P G, Sedensky M M, Meneely P M, Cascorbi H F. Anesthesiology. 1988;69:246–251. doi: 10.1097/00000542-198808000-00015. [DOI] [PubMed] [Google Scholar]
- 10.Morgan P G, Sedensky M M. Anesthesiology. 1994;81:888–898. doi: 10.1097/00000542-199410000-00016. [DOI] [PubMed] [Google Scholar]
- 11.Gamo S, Ogaki M, Nakashima-Tanaka E. Anesthesiology. 1981;54:289–293. doi: 10.1097/00000542-198104000-00006. [DOI] [PubMed] [Google Scholar]
- 12.Krishnan K S, Nash H A. Proc Natl Acad Sci USA. 1990;87:8632–8636. doi: 10.1073/pnas.87.21.8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Campbell D B, Nash H. Proc Natl Acad Sci USA. 1994;91:2135–2139. doi: 10.1073/pnas.91.6.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Simpson V J, Johnson T E. Int Rev Neurobiol. 1996;39:223–241. doi: 10.1016/s0074-7742(08)60668-6. [DOI] [PubMed] [Google Scholar]
- 15.Kendig J J, MacIver M B, Roth S H. Ann NY Acad Sci. 1991;625:37–53. doi: 10.1111/j.1749-6632.1991.tb33828.x. [DOI] [PubMed] [Google Scholar]
- 16.Sedensky M M, Cascorbi H F, Meinwald J, Radford P, Morgan P G. Proc Natl Acad Sci USA. 1994;91:10054–10058. doi: 10.1073/pnas.91.21.10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams B D, Schrank B, Huynh C, Shownkeen R, Waterston R H. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fire A. EMBO J. 1986;5(10):2673–2680. doi: 10.1002/j.1460-2075.1986.tb04550.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blumenthal T, Steward K. In: C. elegans II. Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 117–145. [Google Scholar]
- 23.Waud D R. J Pharmacol Exp Ther. 1972;183:577–607. [PubMed] [Google Scholar]
- 24.Snedecor G W, Cochran W G. Statistical Methods. 6th Ed. Ames: Iowa State Univ. Press; 1968. pp. 271–275. [Google Scholar]
- 25.Park E C, Horvitz H R. Genetics. 1986;113:821–852. doi: 10.1093/genetics/113.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart G W, Argent A C, Dash B C J. Biochim Biophys Acta. 1993;1225:15–25. doi: 10.1016/0925-4439(93)90116-i. [DOI] [PubMed] [Google Scholar]
- 27.Hidaka H, Okazaki K. Neurosci Res. 1993;16(2):73–77. doi: 10.1016/0168-0102(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 28.Hecht R M, Norman T V, Jones W. Genome. 1996;39:459–464. doi: 10.1139/g96-058. [DOI] [PubMed] [Google Scholar]
- 29.Stewart G W, Hepworth-Jones B E, Keen J N, Dash B C, Argent A C, Casimir C M. Blood. 1992;79:1593–1601. [PubMed] [Google Scholar]
- 30.Salzer U, Ahorn H, Prohaska R. Biochim Biophys Acta. 1993;1151:149–152. doi: 10.1016/0005-2736(93)90098-k. [DOI] [PubMed] [Google Scholar]
- 31.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 32.Huang M, Gu G, Ferguson E, Chalfie M. Nature (London) 1995;378:292–295. doi: 10.1038/378292a0. [DOI] [PubMed] [Google Scholar]
- 33.Barnes T, Jin Y, Horvitz H R, Ruvkun G, Hekimi S. J Neurochem. 1996;67:46–57. doi: 10.1046/j.1471-4159.1996.67010046.x. [DOI] [PubMed] [Google Scholar]
- 34.Chalfie M, Sulston J. Dev Biol. 1981;82:358–370. doi: 10.1016/0012-1606(81)90459-0. [DOI] [PubMed] [Google Scholar]
- 35.Canessa C M, Schild L, Buell G, Thorens B, Gautschi I, Horisberger J D, Rossier B C. Nature (London) 1994;367:463–467. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 36.Shreffler W, Wolinsky E. Behav Genet. 1997;27:211–221. doi: 10.1023/a:1025605929373. [DOI] [PubMed] [Google Scholar]
- 37.Foley M, Tilley L. Int J Parasitol. 1997;27:231–240. doi: 10.1016/s0020-7519(96)00152-x. [DOI] [PubMed] [Google Scholar]
- 38.Starich T A, Lee R Y, Panzarella C, Avery L, Shaw J E. J Cell Biol. 1996;134:537–548. doi: 10.1083/jcb.134.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnes T M, Hekimi S. J Neurochem. 1997;69:2251–2260. doi: 10.1046/j.1471-4159.1997.69062251.x. [DOI] [PubMed] [Google Scholar]
- 40.Franks N P, Lieb W R. Nature (London) 1982;300:487–492. doi: 10.1038/300487a0. [DOI] [PubMed] [Google Scholar]