Abstract
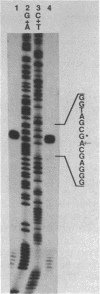
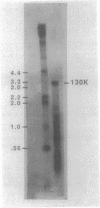
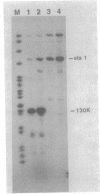
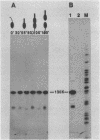
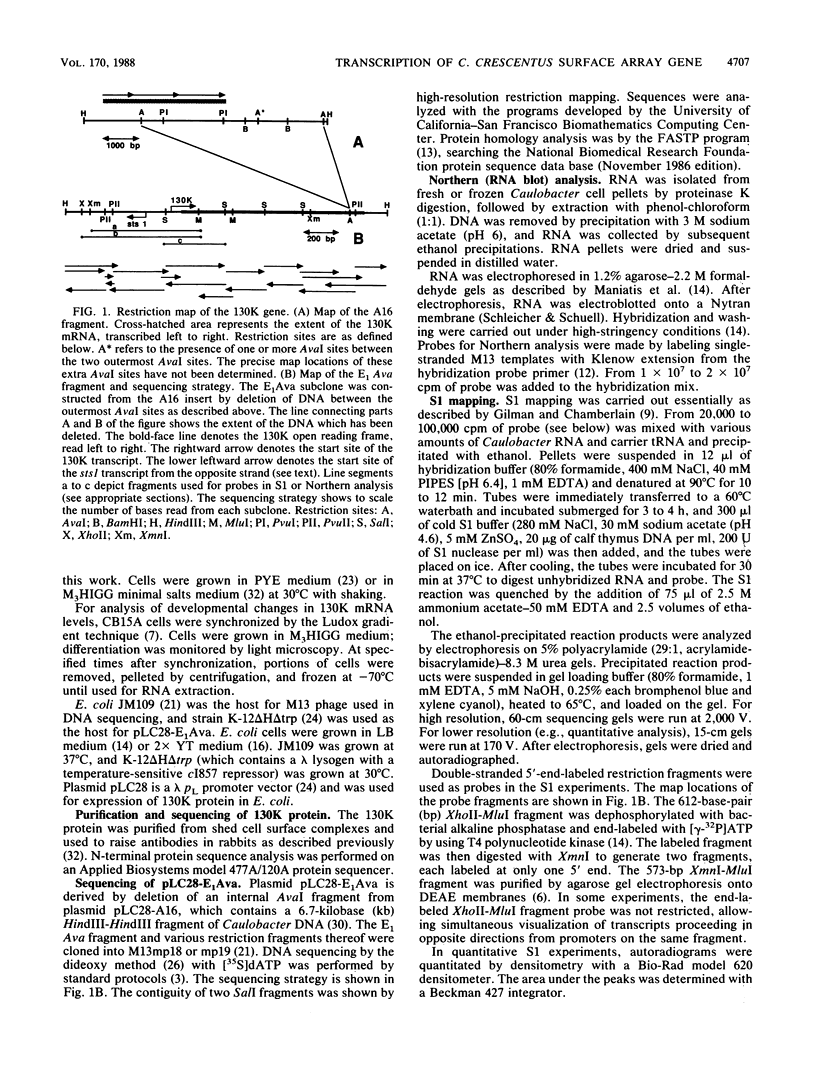
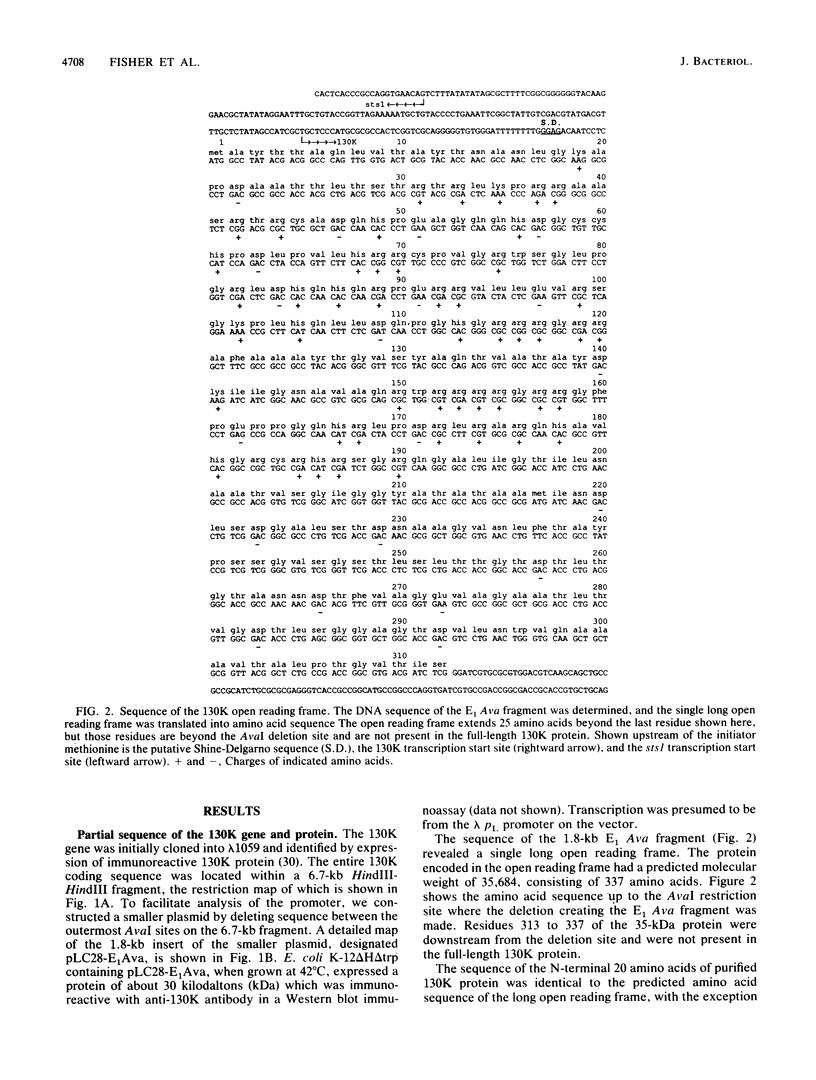
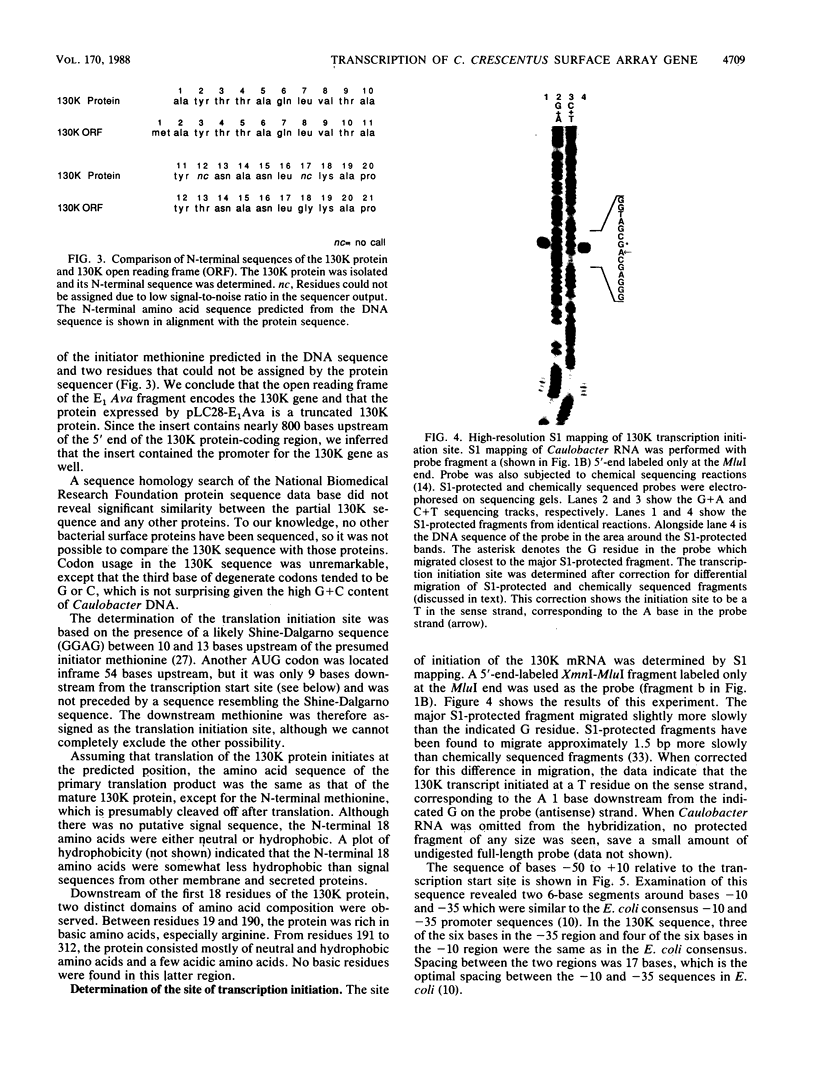
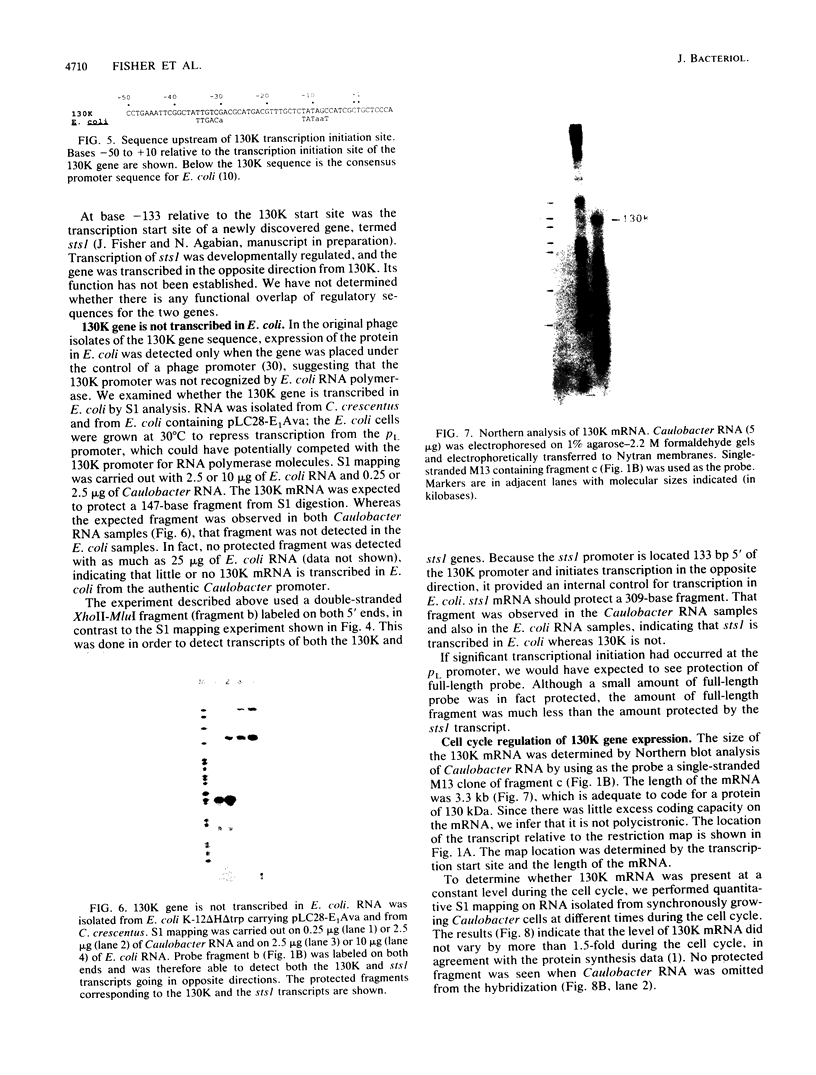
The major component of the paracrystalline surface array of Caulobacter crescentus CB15 and one of the most abundant cellular proteins is a protein designated 130K. We have determined the DNA sequence of the 5' portion of the 130K gene, including the N-terminal one-third of the protein coding region, and analyzed the transcription of the gene. The site of transcription initiation was determined by S1 mapping of Caulobacter RNA. Although the DNA sequence upstream from the transcription start site showed significant homology to the consensus promoter sequences of Escherichia coli, S1 analysis of RNA from E. coli carrying the 130K gene on a plasmid indicated that the 130K promoter was not transcribed by E. coli RNA polymerase in vivo. Quantitative S1 analysis of RNA isolated from synchronously growing Caulobacter cells suggested that this promoter was not under developmental regulation; the amount of 130K transcript varied no more than 1.5-fold during the cell cycle. The length of the 130K mRNA was determined to be 3.3 kilobases by Northern (RNA blot) analysis, indicating that the 130K mRNA is not part of a polycistron. The amino acid sequence predicted from the DNA sequence agreed well with the N-terminal amino acid sequence determined by sequencing of the 130K protein. The 130K protein appears to be synthesized without an N-terminal leader sequence, but the N-terminal 20 amino acids are relatively hydrophobic and may function like a signal sequence during transmembrane translocation.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agabian N., Evinger M., Parker G. Generation of asymmetry during development. Segregation of type-specific proteins in Caulobacter. J Cell Biol. 1979 Apr;81(1):123–136. doi: 10.1083/jcb.81.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya K., Bellofatto V., Shapiro L., Feingold J. Transcription initiation in vitro and in vivo at a highly conserved promoter within a 16 S ribosomal RNA gene. J Mol Biol. 1986 Jan 5;187(1):1–14. doi: 10.1016/0022-2836(86)90401-8. [DOI] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. S., Mullin D., Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci U S A. 1986 May;83(9):2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi R. H., Wang L. F. Multiple procaryotic ribonucleic acid polymerase sigma factors. Microbiol Rev. 1986 Sep;50(3):227–243. doi: 10.1128/mr.50.3.227-243.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Evinger M., Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977 Oct;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P. R., Agabian N. The nucleotide sequence of the Mr = 28,500 flagellin gene of Caulobacter crescentus. J Biol Chem. 1983 Jun 25;258(12):7395–7401. [PubMed] [Google Scholar]
- Gilman M. Z., Chamberlin M. J. Developmental and genetic regulation of Bacillus subtilis genes transcribed by sigma 28-RNA polymerase. Cell. 1983 Nov;35(1):285–293. doi: 10.1016/0092-8674(83)90231-3. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Chamberlin M. J. DNA sequence analysis suggests that expression of flagellar and chemotaxis genes in Escherichia coli and Salmonella typhimurium is controlled by an alternative sigma factor. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6422–6424. doi: 10.1073/pnas.84.18.6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Lechner J., Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J Biol Chem. 1987 Jul 15;262(20):9724–9729. [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983 Apr 14;302(5909):630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Gill P. R., Parker G., Agabian N. Cloning of developmentally regulated flagellin genes from Caulobacter crescentus via immunoprecipitation of polyribosomes. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6847–6851. doi: 10.1073/pnas.79.22.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich S. A., Newton A. Promoter mapping and cell cycle regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullin D., Minnich S., Chen L. S., Newton A. A set of positively regulated flagellar gene promoters in Caulobacter crescentus with sequence homology to the nif gene promoters of Klebsiella pneumoniae. J Mol Biol. 1987 Jun 20;195(4):939–943. doi: 10.1016/0022-2836(87)90497-9. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohta N., Chen L. S., Swanson E., Newton A. Transcriptional regulation of a periodically controlled flagellar gene operon in Caulobacter crescentus. J Mol Biol. 1985 Nov 5;186(1):107–115. doi: 10.1016/0022-2836(85)90261-x. [DOI] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J., Peters M., Lottspeich F., Schäfer W., Baumeister W. Nucleotide sequence analysis of the gene encoding the Deinococcus radiodurans surface protein, derived amino acid sequence, and complementary protein chemical studies. J Bacteriol. 1987 Nov;169(11):5216–5223. doi: 10.1128/jb.169.11.5216-5223.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remaut E., Stanssens P., Fiers W. Plasmid vectors for high-efficiency expression controlled by the PL promoter of coliphage lambda. Gene. 1981 Oct;15(1):81–93. doi: 10.1016/0378-1119(81)90106-2. [DOI] [PubMed] [Google Scholar]
- Ross C. M., Winkler M. E. Structure of the Caulobacter crescentus trpFBA operon. J Bacteriol. 1988 Feb;170(2):757–768. doi: 10.1128/jb.170.2.757-768.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. Determinant of cistron specificity in bacterial ribosomes. Nature. 1975 Mar 6;254(5495):34–38. doi: 10.1038/254034a0. [DOI] [PubMed] [Google Scholar]
- Sleytr U. B., Messner P. Crystalline surface layers on bacteria. Annu Rev Microbiol. 1983;37:311–339. doi: 10.1146/annurev.mi.37.100183.001523. [DOI] [PubMed] [Google Scholar]
- Smit J., Agabian N. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J Cell Biol. 1982 Oct;95(1):41–49. doi: 10.1083/jcb.95.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Agabian N. Cloning of the major protein of the Caulobacter crescentus periodic surface layer: detection and characterization of the cloned peptide by protein expression assays. J Bacteriol. 1984 Dec;160(3):1137–1145. doi: 10.1128/jb.160.3.1137-1145.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J., Grano D. A., Glaeser R. M., Agabian N. Periodic surface array in Caulobacter crescentus: fine structure and chemical analysis. J Bacteriol. 1981 Jun;146(3):1135–1150. doi: 10.1128/jb.146.3.1135-1150.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Watson M. E. Compilation of published signal sequences. Nucleic Acids Res. 1984 Jul 11;12(13):5145–5164. doi: 10.1093/nar/12.13.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. T., Lodish H. F. Multiple mechanisms of protein insertion into and across membranes. Science. 1985 Oct 25;230(4724):400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Winkler M. E., Schoenlein P. V., Ross C. M., Barrett J. T., Ely B. Genetic and physical analyses of Caulobacter crescentus trp genes. J Bacteriol. 1984 Oct;160(1):279–287. doi: 10.1128/jb.160.1.279-287.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata H., Adachi T., Tsuboi A., Takao M., Sasaki T., Tsukagoshi N., Udaka S. Cloning and characterization of the 5' region of the cell wall protein gene operon in Bacillus brevis 47. J Bacteriol. 1987 Mar;169(3):1239–1245. doi: 10.1128/jb.169.3.1239-1245.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]