Abstract
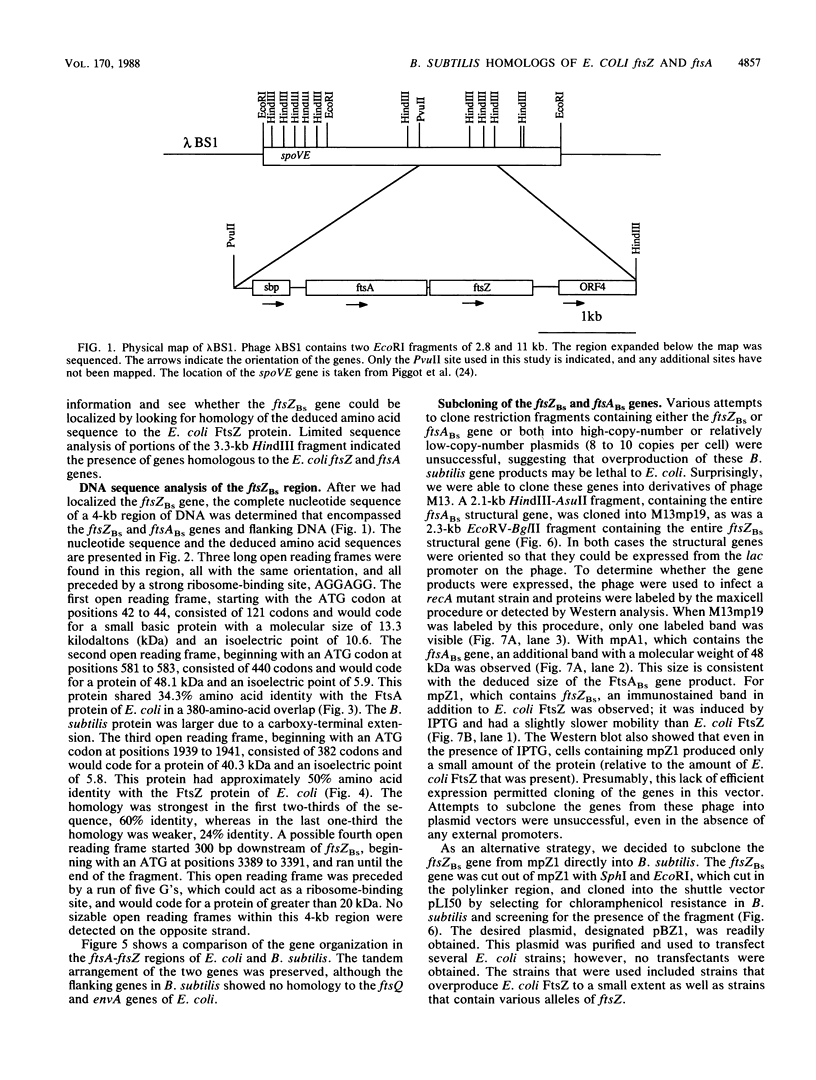
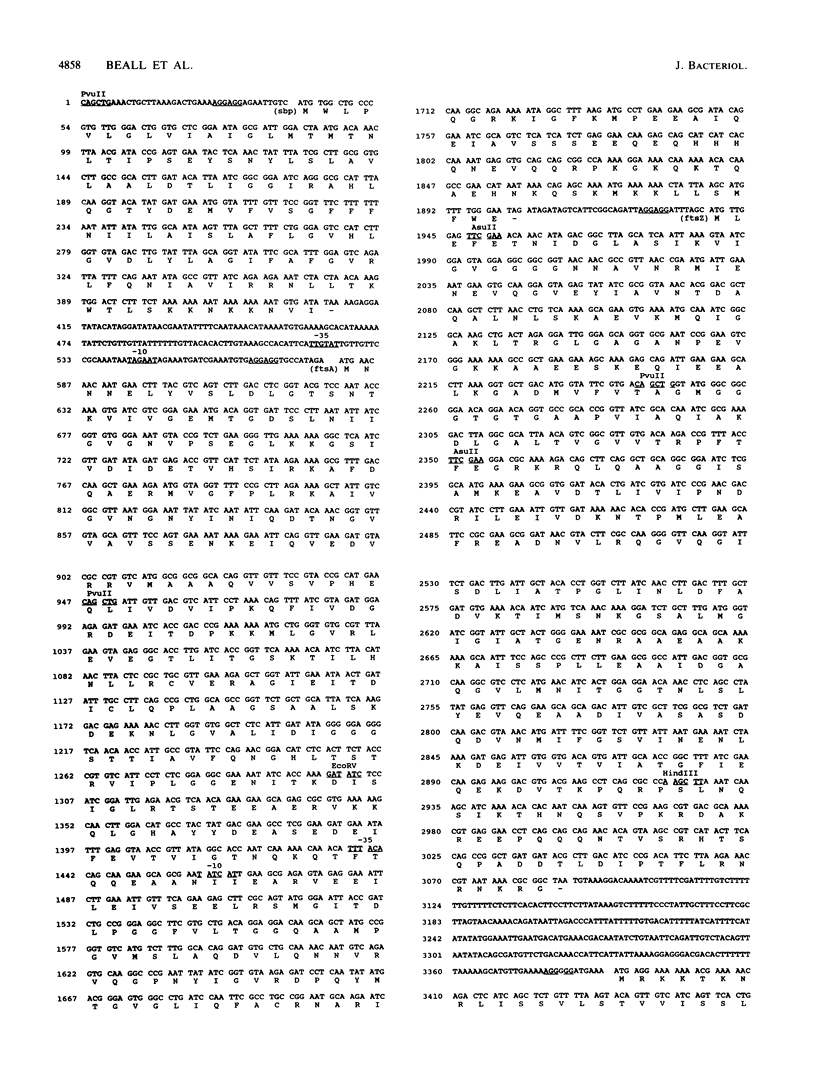
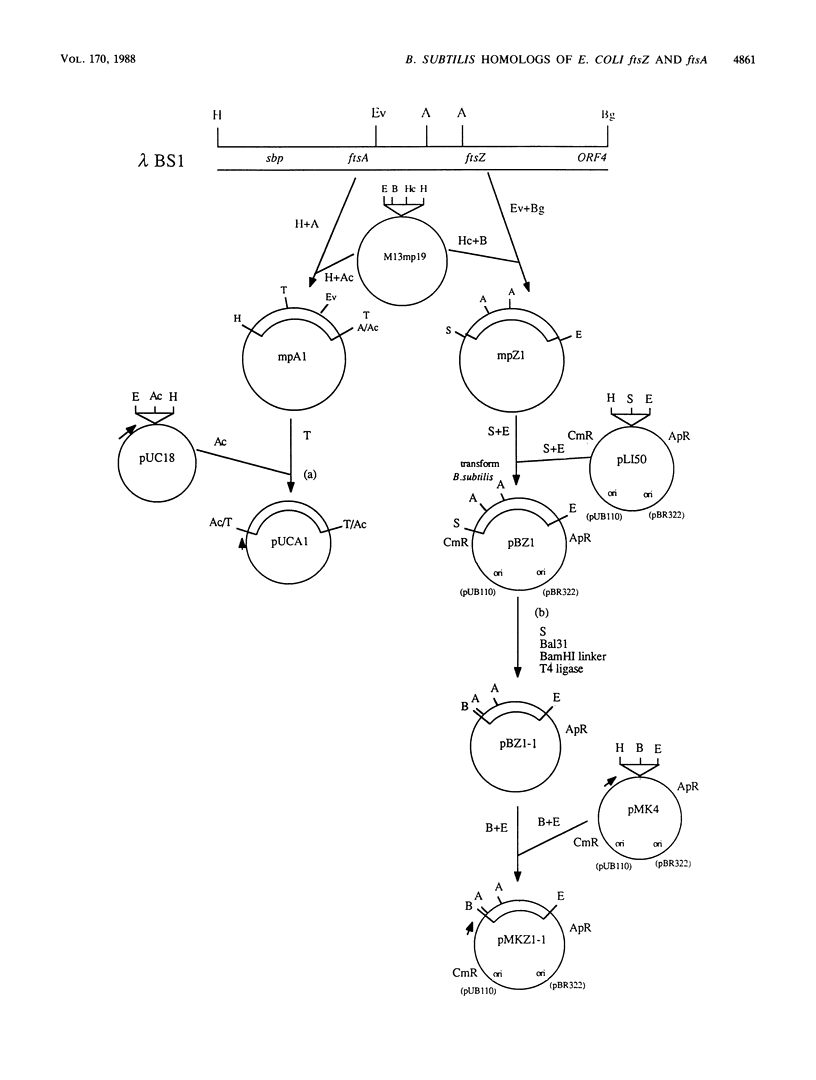
The Bacillus subtilis homolog of the Escherichia coli ftsZ gene was isolated by screening a B. subtilis genomic library with anti-E. coli FtsZ antiserum. DNA sequence analysis of a 4-kilobase region revealed three open reading frames. One of these coded for a protein that was about 50% homologous to the E. coli FtsZ protein. The open reading frame just upstream of ftsZ coded for a protein that was 34% homologous to the E. coli FtsA protein. The open reading frames flanking these two B. subtilis genes showed no relationship to those found in E. coli. Expression of the B. subtilis ftsZ and ftsA genes in E. coli was lethal, since neither of these genes could be cloned on plasmid vectors unless promoter sequences were first removed. Cloning the B. subtilis ftsZ gene under the control of the lac promoter resulted in an IPTGs phenotype that could be suppressed by overproduction of E. coli FtsZ. These genes mapped at 135 degrees on the B. subtilis genetic map near previously identified cell division mutations.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrup L., Atlung T., Ogasawara N., Yoshikawa H., Hansen F. G. Interaction of the Bacillus subtilis DnaA-like protein with the Escherichia coli DnaA protein. J Bacteriol. 1988 Mar;170(3):1333–1338. doi: 10.1128/jb.170.3.1333-1338.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister H., Wake R. G. Characterization and mapping of temperature-sensitive division initiation mutations of Bacillus subtilis. J Bacteriol. 1981 Feb;145(2):1042–1051. doi: 10.1128/jb.145.2.1042-1051.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Copeland J. C., Marmur J. Identification of conserved genetic functions in Bacillus by use of temperature-sensitive mutants. Bacteriol Rev. 1968 Dec;32(4 Pt 1):302–312. [PMC free article] [PubMed] [Google Scholar]
- Corton J. C., Ward J. E., Jr, Lutkenhaus J. Analysis of cell division gene ftsZ (sulB) from gram-negative and gram-positive bacteria. J Bacteriol. 1987 Jan;169(1):1–7. doi: 10.1128/jb.169.1.1-7.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedonder R. A., Lepesant J. A., Lepesant-Kejzlarová J., Billault A., Steinmetz M., Kunst F. Construction of a kit of reference strains for rapid genetic mapping in Bacillus subtilis 168. Appl Environ Microbiol. 1977 Apr;33(4):989–993. doi: 10.1128/aem.33.4.989-993.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau D., Davidoff-Abelson R., Scher B., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: phenotypic characterization of radiation-sensitive recombination-deficient mutants. J Bacteriol. 1973 Apr;114(1):273–286. doi: 10.1128/jb.114.1.273-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari E., Henner D. J., Hoch J. A. Isolation of Bacillus subtilis genes from a charon 4A library. J Bacteriol. 1981 Apr;146(1):430–432. doi: 10.1128/jb.146.1.430-432.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari F. A., Nguyen A., Lang D., Hoch J. A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983 Jun;154(3):1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981 Apr 30;290(5809):797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Jones C., Holland I. B. Role of the SulB (FtsZ) protein in division inhibition during the SOS response in Escherichia coli: FtsZ stabilizes the inhibitor SulA in maxicells. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6045–6049. doi: 10.1073/pnas.82.18.6045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Love P. E., Yasbin R. E. Genetic characterization of the inducible SOS-like system of Bacillus subtilis. J Bacteriol. 1984 Dec;160(3):910–920. doi: 10.1128/jb.160.3.910-920.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F. Coupling of DNA replication and cell division: sulB is an allele of ftsZ. J Bacteriol. 1983 Jun;154(3):1339–1346. doi: 10.1128/jb.154.3.1339-1346.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. F., Wolf-Watz H., Donachie W. D. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol. 1980 May;142(2):615–620. doi: 10.1128/jb.142.2.615-620.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Nukushina J. I., Ikeda Y. Genetic analysis of the developmental processes during germination and outgrowth of Bacillus subtilis spores with temperature-sensitive mutants. Genetics. 1969 Sep;63(1):63–74. doi: 10.1093/genetics/63.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara N., Moriya S., von Meyenburg K., Hansen F. G., Yoshikawa H. Conservation of genes and their organization in the chromosomal replication origin region of Bacillus subtilis and Escherichia coli. EMBO J. 1985 Dec 1;4(12):3345–3350. doi: 10.1002/j.1460-2075.1985.tb04087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott R. W., Goodman L. E., Ganesan A. T. Expression of the Bacillus subtilis polC gene in Escherichia coli. Mol Gen Genet. 1987 May;207(2-3):335–341. doi: 10.1007/BF00331598. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Chak K. F., Bugaichuk U. D. Isolation and characterization of a clone of the spoVE locus of Bacillus subtilis. J Gen Microbiol. 1986 Jul;132(7):1875–1881. doi: 10.1099/00221287-132-7-1875. [DOI] [PubMed] [Google Scholar]
- Piggot P. J., Hoch J. A. Revised genetic linkage map of Bacillus subtilis. Microbiol Rev. 1985 Jun;49(2):158–179. doi: 10.1128/mr.49.2.158-179.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard M., Hirota Y. Process of cellular division in Escherichia coli: physiological study on thermosensitive mutants defective in cell division. J Bacteriol. 1973 Oct;116(1):314–322. doi: 10.1128/jb.116.1.314-322.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancar A., Hack A. M., Rupp W. D. Simple method for identification of plasmid-coded proteins. J Bacteriol. 1979 Jan;137(1):692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. A., Yasbin R. E., Young F. E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984 Jul-Aug;29(1-2):21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- Swanberg S. L., Wang J. C. Cloning and sequencing of the Escherichia coli gyrA gene coding for the A subunit of DNA gyrase. J Mol Biol. 1987 Oct 20;197(4):729–736. doi: 10.1016/0022-2836(87)90479-7. [DOI] [PubMed] [Google Scholar]
- Van Alstyne D., Simon M. I. Division mutants of Bacillus subtilis: isolation and PBS1 transduction of division-specific markers. J Bacteriol. 1971 Dec;108(3):1366–1379. doi: 10.1128/jb.108.3.1366-1379.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeyar M. A., Zahler S. A. Chromosomal insertions of Tn917 in Bacillus subtilis. J Bacteriol. 1986 Aug;167(2):530–534. doi: 10.1128/jb.167.2.530-534.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. F. A lacZ-ftsZ gene fusion is an analog of the cell division inhibitor sulA. J Bacteriol. 1984 Mar;157(3):815–820. doi: 10.1128/jb.157.3.815-820.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985 Oct;42(3):941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Lutkenhaus J. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene. 1985;36(3):241–247. doi: 10.1016/0378-1119(85)90179-9. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]
- Young M. Genetic mapping of sporulation operons in Bacillus subtilis using a thermosensitive sporulation mutant. J Bacteriol. 1975 Jun;122(3):1109–1116. doi: 10.1128/jb.122.3.1109-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]



