Abstract
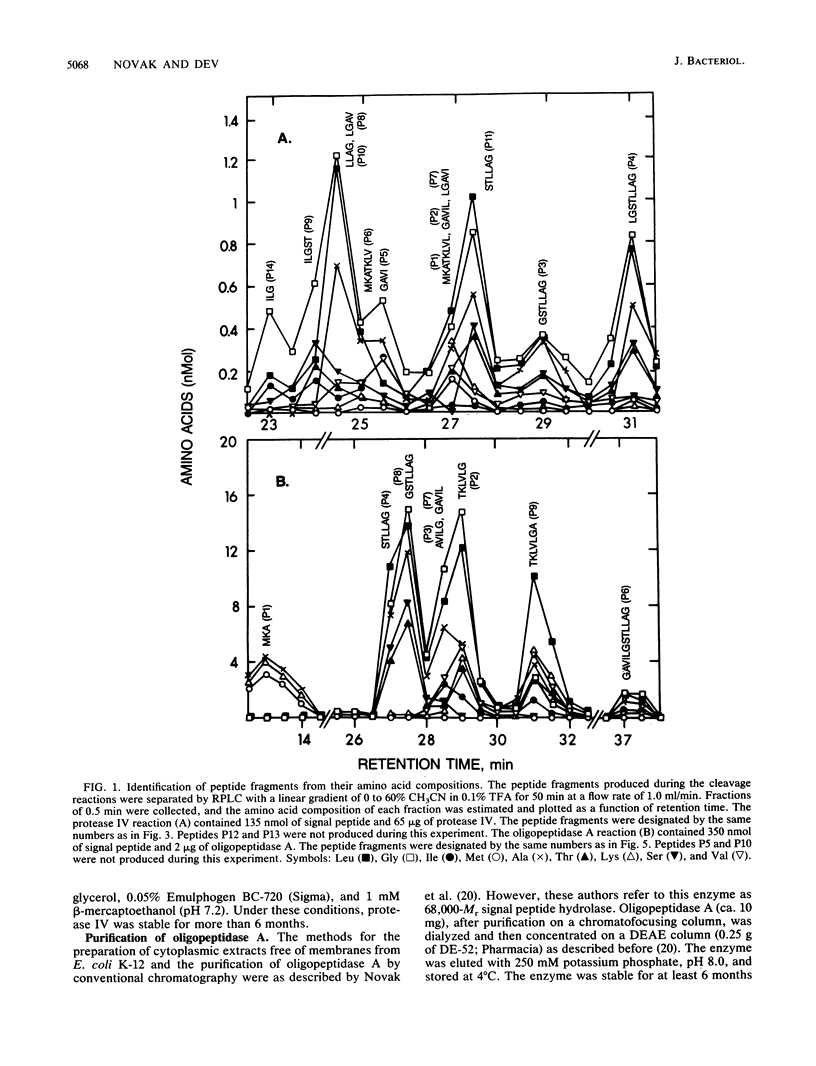
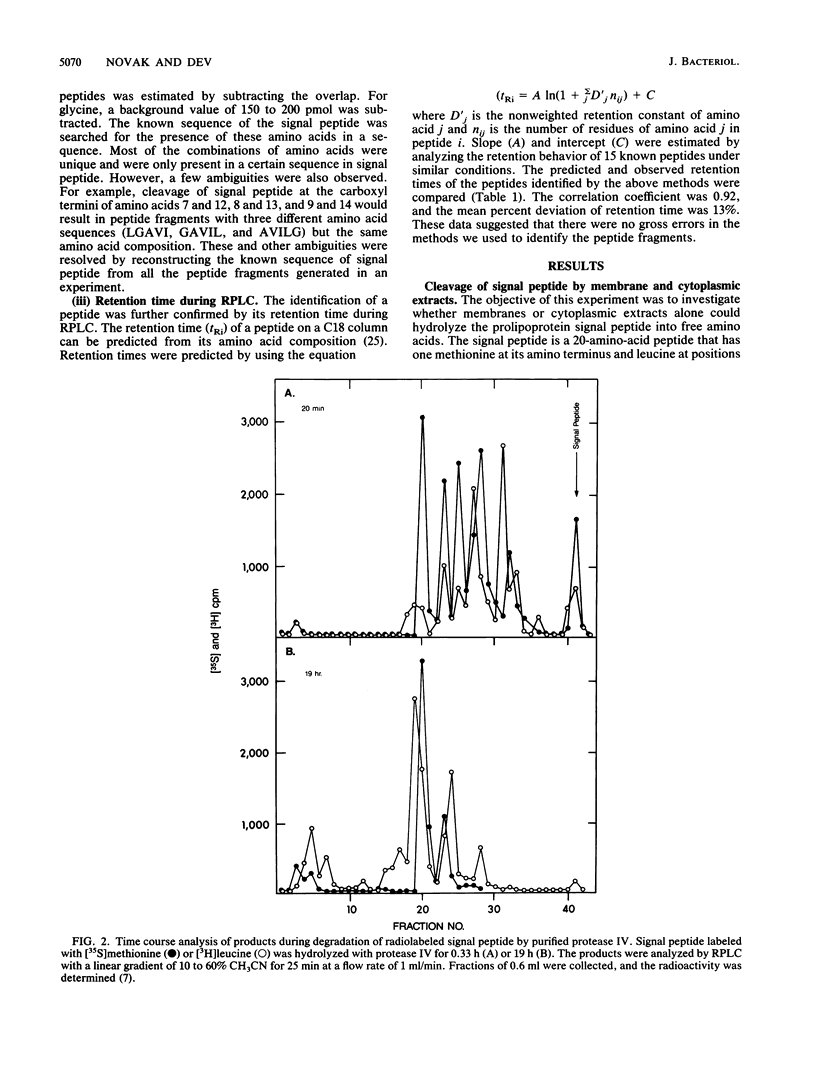
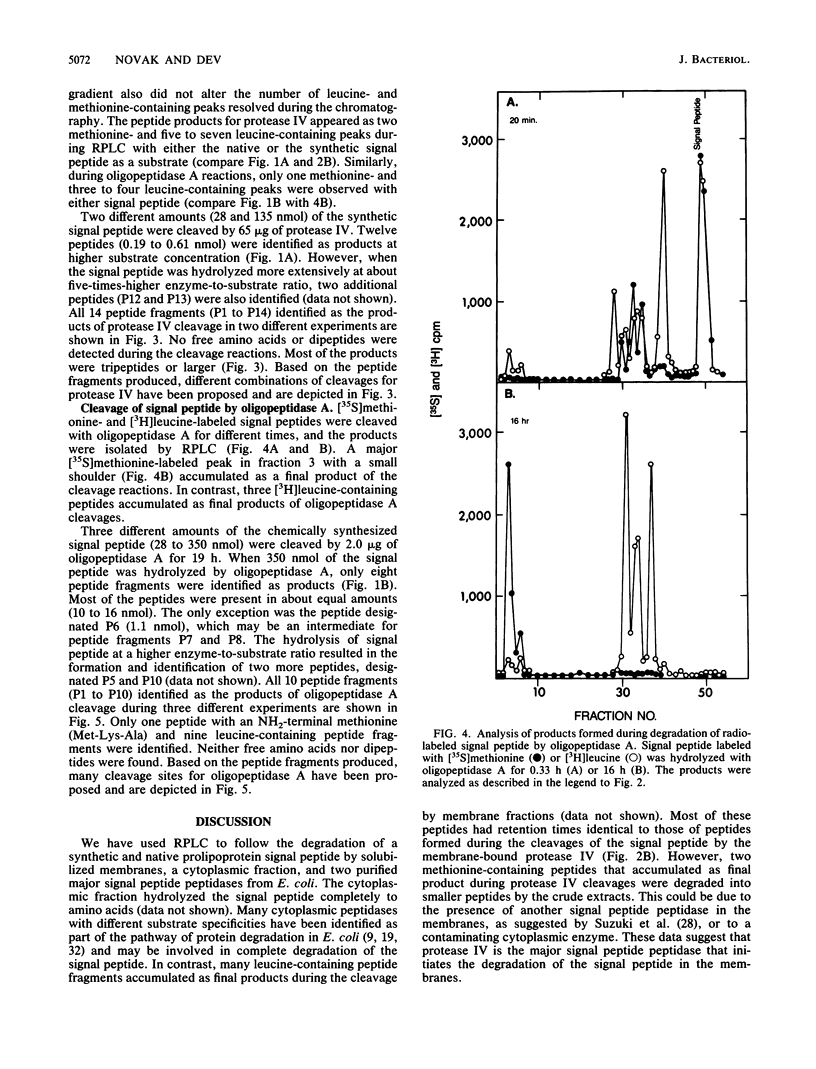
The degradation of the prolipoprotein signal peptide in vitro by membranes, cytoplasmic fraction, and two purified major signal peptide peptidases from Escherichia coli was followed by reverse-phase liquid chromatography (RPLC). The cytoplasmic fraction hydrolyzed the signal peptide completely into amino acids. In contrast, many peptide fragments accumulated as final products during the cleavage by a membrane fraction. Most of the peptides were similar to the peptides formed during the cleavage of the signal peptide by the purified membrane-bound signal peptide peptidase, protease IV. Peptide fragments generated during the cleavage of the signal peptide by protease IV and a cytoplasmic enzyme, oligopeptidase A, were identified from their amino acid compositions, their retention times during RPLC, and knowledge of the amino acid sequence of the signal peptide. Both enzymes were endopeptidases, as neither dipeptides nor free amino acids were formed during the cleavage reactions. Protease IV cleaved the signal peptide predominantly in the hydrophobic segment (residues 7 to 14). Protease IV required substrates with hydrophobic amino acids at the primary and the adjacent substrate-binding sites, with a minimum of three amino acids on either side of the scissile bond. Oligopeptidase A cleaved peptides (minimally five residues) that had either alanine or glycine at the P'1 (primary binding site) or at the P1 (preceding P'1) site of the substrate. These results support the hypothesis that protease IV is the major signal peptide peptidase in membranes that initiates the degradation of the signal peptide by making endoproteolytic cuts; oligopeptidase A and other cytoplasmic enzymes further degrade the partially degraded portions of the signal peptide that may be diffused or transported back into the cytoplasm from the membranes.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austen B. M., Hermon-Taylor J., Kaderbhai M. A., Ridd D. H. Design and synthesis of a consensus signal sequence that inhibits protein translocation into rough microsomal vesicles. Biochem J. 1984 Nov 15;224(1):317–325. doi: 10.1042/bj2240317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson S. A., Hall M. N., Silhavy T. J. Genetic analysis of protein export in Escherichia coli K12. Annu Rev Biochem. 1985;54:101–134. doi: 10.1146/annurev.bi.54.070185.000533. [DOI] [PubMed] [Google Scholar]
- Bidlingmeyer B. A., Cohen S. A., Tarvin T. L. Rapid analysis of amino acids using pre-column derivatization. J Chromatogr. 1984 Dec 7;336(1):93–104. doi: 10.1016/s0378-4347(00)85133-6. [DOI] [PubMed] [Google Scholar]
- Chen L., Tai P. C., Briggs M. S., Gierasch L. M. Protein translocation into Escherichia coli membrane vesicles is inhibited by functional synthetic signal peptides. J Biol Chem. 1987 Feb 5;262(4):1427–1429. [PubMed] [Google Scholar]
- Chung C. H., Goldberg A. L. Purification and characterization of protease So, a cytoplasmic serine protease in Escherichia coli. J Bacteriol. 1983 Apr;154(1):231–238. doi: 10.1128/jb.154.1.231-238.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett R. J., Roche R. S. The identification of large peptide fragments produced from proteins of known sequence: a computerized approach using amino acid composition indexes and its application to thermolysin. Anal Biochem. 1987 May 1;162(2):546–552. doi: 10.1016/0003-2697(87)90432-5. [DOI] [PubMed] [Google Scholar]
- Dev I. K., Ray P. H. Rapid assay and purification of a unique signal peptidase that processes the prolipoprotein from Escherichia coli B. J Biol Chem. 1984 Sep 10;259(17):11114–11120. [PubMed] [Google Scholar]
- Goldberg A. L., Swamy K. H., Chung C. H., Larimore F. S. Proteases in Escherichia coli. Methods Enzymol. 1981;80(Pt 100):680–702. doi: 10.1016/s0076-6879(81)80052-3. [DOI] [PubMed] [Google Scholar]
- Habener J. F., Rosenblatt M., Dee P. C., Potts J. T., Jr Cellular processing of pre-proparathyroid hormone involves rapid hydrolysis of the leader sequence. J Biol Chem. 1979 Nov 10;254(21):10596–10599. [PubMed] [Google Scholar]
- Heinrikson R. L., Meredith S. C. Amino acid analysis by reverse-phase high-performance liquid chromatography: precolumn derivatization with phenylisothiocyanate. Anal Biochem. 1984 Jan;136(1):65–74. doi: 10.1016/0003-2697(84)90307-5. [DOI] [PubMed] [Google Scholar]
- Hussain M., Ozawa Y., Ichihara S., Mizushima S. Signal peptide digestion in Escherichia coli. Effect of protease inhibitors on hydrolysis of the cleaved signal peptide of the major outer-membrane lipoprotein. Eur J Biochem. 1982 Dec;129(1):233–239. doi: 10.1111/j.1432-1033.1982.tb07044.x. [DOI] [PubMed] [Google Scholar]
- Ichihara S., Beppu N., Mizushima S. Protease IV, a cytoplasmic membrane protein of Escherichia coli, has signal peptide peptidase activity. J Biol Chem. 1984 Aug 10;259(15):9853–9857. [PubMed] [Google Scholar]
- Ichihara S., Suzuki T., Suzuki M., Mizushima S. Molecular cloning and sequencing of the sppA gene and characterization of the encoded protease IV, a signal peptide peptidase, of Escherichia coli. J Biol Chem. 1986 Jul 15;261(20):9405–9411. [PubMed] [Google Scholar]
- Inouye S., Wang S., Sekizawa J., Halegoua S., Inouye M. Amino acid sequence for the peptide extension on the prolipoprotein of the Escherichia coli outer membrane. Proc Natl Acad Sci U S A. 1977 Mar;74(3):1004–1008. doi: 10.1073/pnas.74.3.1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai M., Inouye M. Association of the prolipoprotein accumulated in the presence of globomycin with the outer membrane of Escherichia coli. Eur J Biochem. 1983 Jan 17;130(1):27–32. doi: 10.1111/j.1432-1033.1983.tb07112.x. [DOI] [PubMed] [Google Scholar]
- Jackson R. C., Blobel G. Post-translational cleavage of presecretory proteins with an extract of rough microsomes from dog pancreas containing signal peptidase activity. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5598–5602. doi: 10.1073/pnas.74.12.5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren R., Burstein Y., Soreq H. Synthetic leader peptide modulates secretion of proteins from microinjected Xenopus oocytes. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7205–7209. doi: 10.1073/pnas.80.23.7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P., Ray P. H., Dev I. K. Localization and purification of two enzymes from Escherichia coli capable of hydrolyzing a signal peptide. J Biol Chem. 1986 Jan 5;261(1):420–427. [PubMed] [Google Scholar]
- Pacaud M. Purification and characterization of two novel proteolytic enzymes in membranes of Escherichia coli. Protease IV and protease V. J Biol Chem. 1982 Apr 25;257(8):4333–4339. [PubMed] [Google Scholar]
- Palmer S. M., St John A. C. Characterization of a membrane-associated serine protease in Escherichia coli. J Bacteriol. 1987 Apr;169(4):1474–1479. doi: 10.1128/jb.169.4.1474-1479.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman D., Halvorson H. O. A putative signal peptidase recognition site and sequence in eukaryotic and prokaryotic signal peptides. J Mol Biol. 1983 Jun 25;167(2):391–409. doi: 10.1016/s0022-2836(83)80341-6. [DOI] [PubMed] [Google Scholar]
- Ray P., Dev I., MacGregor C., Bassford P., Jr Signal peptidases. Curr Top Microbiol Immunol. 1986;125:75–102. doi: 10.1007/978-3-642-71251-7_7. [DOI] [PubMed] [Google Scholar]
- Silhavy T. J., Benson S. A., Emr S. D. Mechanisms of protein localization. Microbiol Rev. 1983 Sep;47(3):313–344. doi: 10.1128/mr.47.3.313-344.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer S. J., Maher P. A., Yaffe M. P. On the translocation of proteins across membranes. Proc Natl Acad Sci U S A. 1987 Feb;84(4):1015–1019. doi: 10.1073/pnas.84.4.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Itoh A., Ichihara S., Mizushima S. Characterization of the sppA gene coding for protease IV, a signal peptide peptidase of Escherichia coli. J Bacteriol. 1987 Jun;169(6):2523–2528. doi: 10.1128/jb.169.6.2523-2528.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimr E. R., Green L., Miller C. G. Oligopeptidase-deficient mutants of Salmonella typhimurium. J Bacteriol. 1983 Mar;153(3):1259–1265. doi: 10.1128/jb.153.3.1259-1265.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W. T., Lodish H. F. Multiple mechanisms of protein insertion into and across membranes. Science. 1985 Oct 25;230(4724):400–407. doi: 10.1126/science.4048938. [DOI] [PubMed] [Google Scholar]
- Wickner W., Moore K., Dibb N., Geissert D., Rice M. Inhibition of purified Escherichia coli leader peptidase by the leader (signal) peptide of bacteriophage M13 procoat. J Bacteriol. 1987 Aug;169(8):3821–3822. doi: 10.1128/jb.169.8.3821-3822.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen C., Green L., Miller C. G. Degradation of intracellular protein in Salmonella typhimurium peptidase mutants. J Mol Biol. 1980 Oct 15;143(1):21–33. doi: 10.1016/0022-2836(80)90122-9. [DOI] [PubMed] [Google Scholar]
- Zwizinski C., Wickner W. Purification and characterization of leader (signal) peptidase from Escherichia coli. J Biol Chem. 1980 Aug 25;255(16):7973–7977. [PubMed] [Google Scholar]


