Abstract
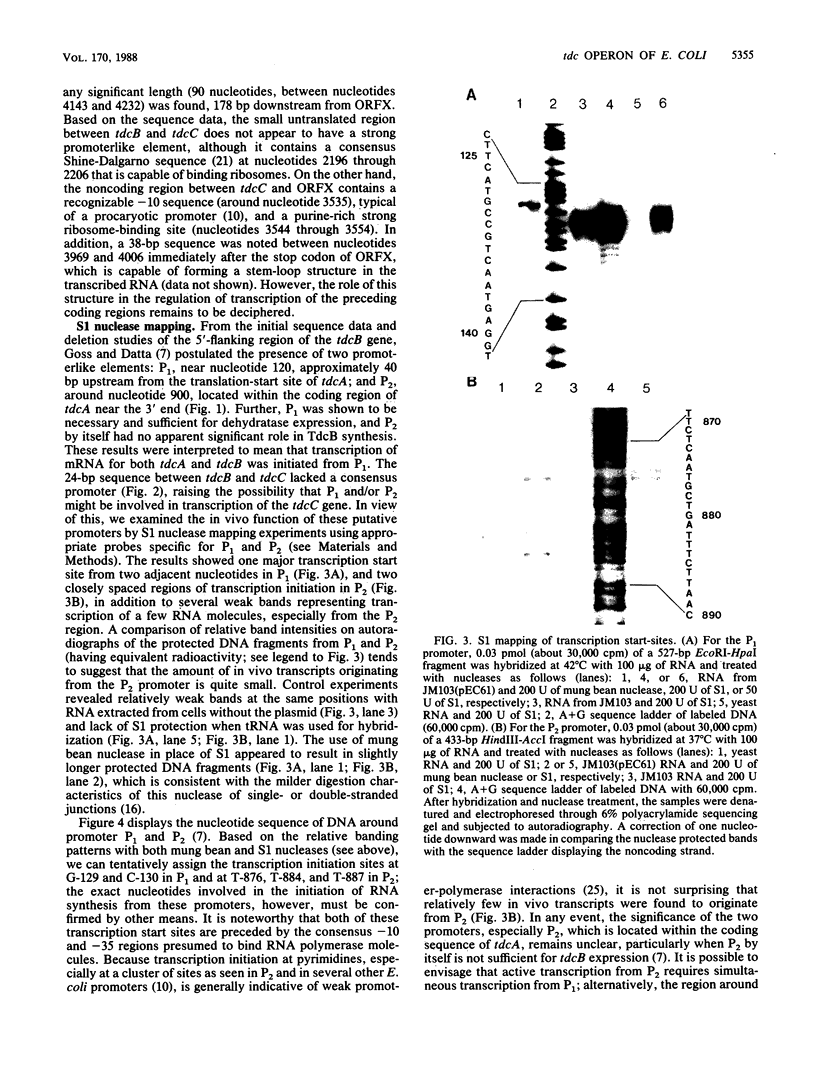
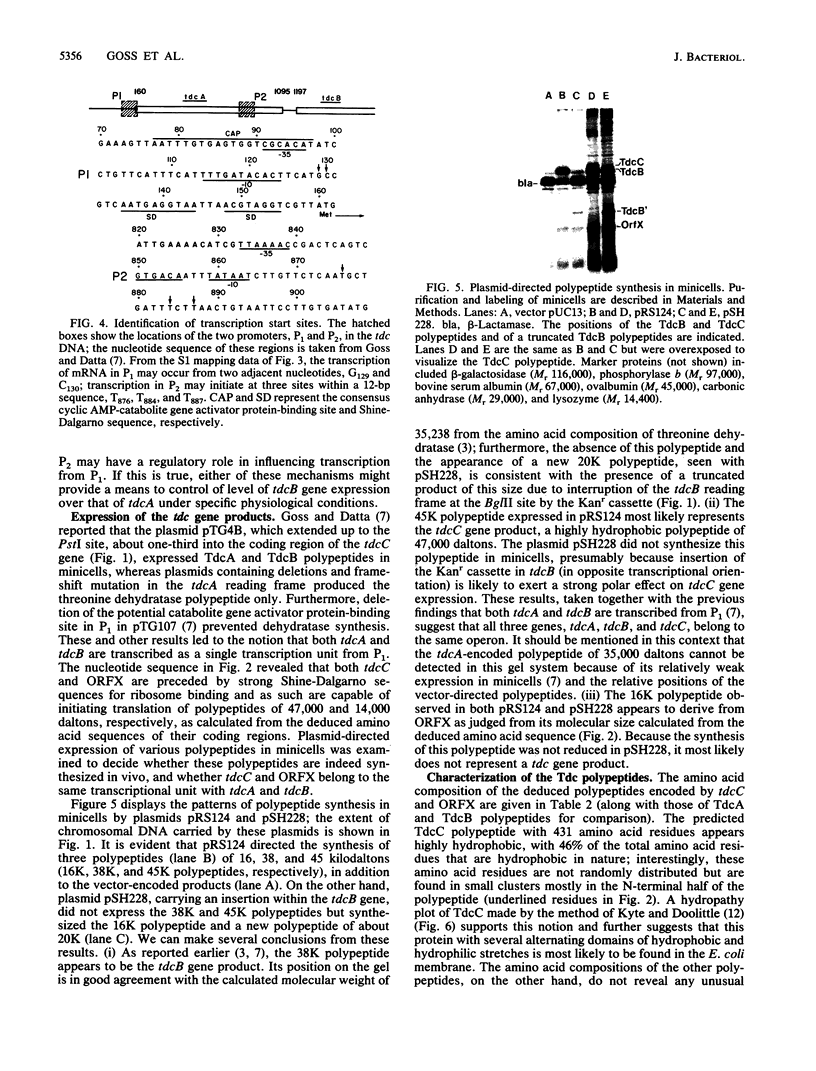
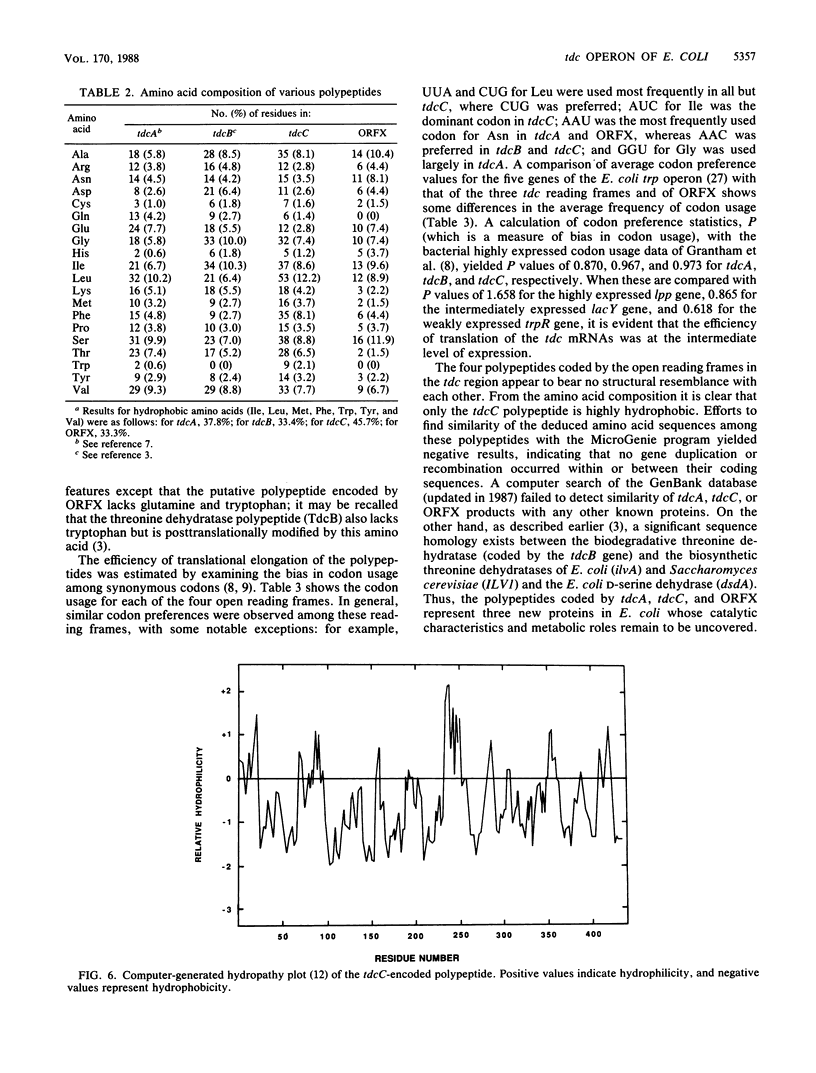
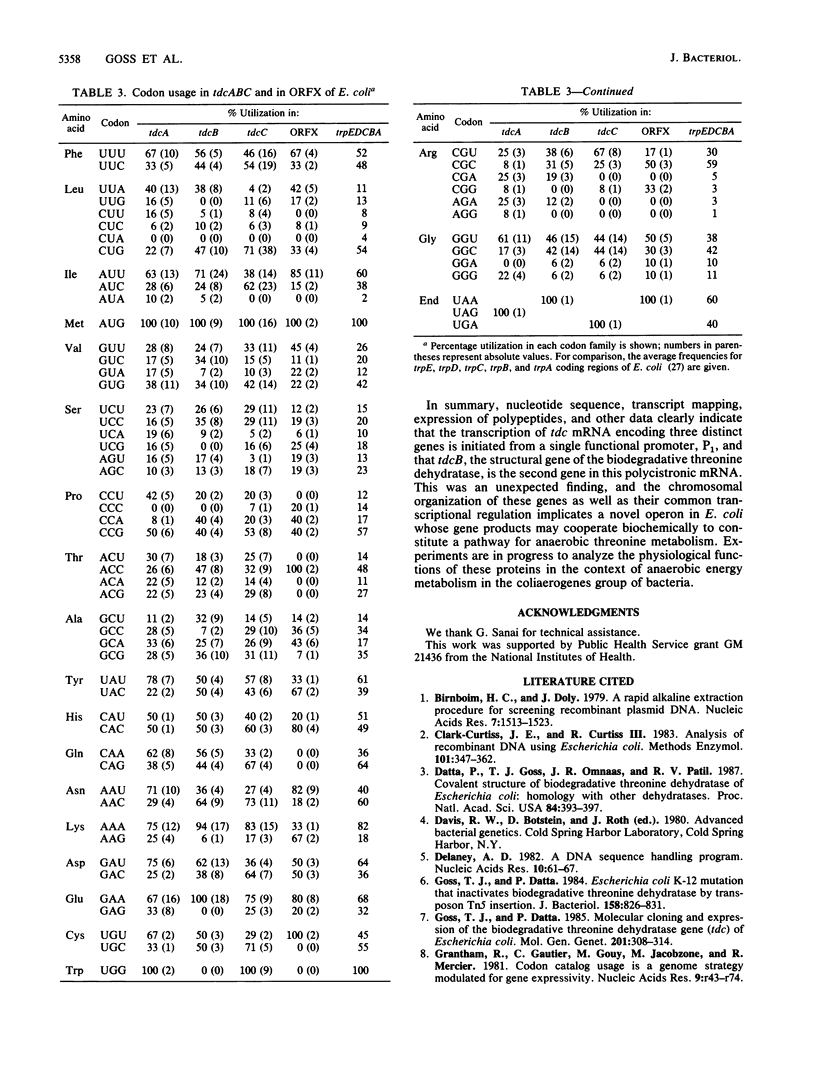
The nucleotide sequence of a 2-kilobase DNA fragment of the tdc region of Escherichia coli K-12, previously cloned in this laboratory, revealed two open reading frames, tdcC and ORFX, downstream from the tdcB gene (formerly designated tdc) encoding biodegradative threonine dehydratase. A 24-base-pair sequence separated tdcC from the dehydratase coding region, and an untranslated region of 60 nucleotides, which contains a recognizable -10 consensus sequence, was found between tdcC and ORFX. The deduced amino acid sequence of tdcC showed it to be a large hydrophobic polypeptide of 431 amino acid residues, whereas ORFX coded for a small 135-residue polypeptide lacking glutamine and tryptophan. A computer-assisted sequence analysis revealed no similarity among the tdcB, tdcC, and ORFX polypeptides, and a search of the GenBank database failed to detect similarity with any other known proteins. The tdc genes and ORFX showed similar codon usage and, in analogy with other bacterial genes, showed codon usage typical for genes expressed at an intermediate level. Transcriptional analysis with S1 nuclease indicated two distinct transcription start sites upstream of the tdcB gene in regions previously identified as promoterlike elements P1 and P2. Interestingly, expression of tdcB and tdcC, but not ORFX, was contingent upon the presence of P1. These results taken together tend to suggest that the biodegradative threonine dehydratase is the second gene in a polycistronic transcription unit constituting a novel operon (tdcABC) in E. coli implicated in anaerobic threonine metabolism.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Curtiss J. E., Curtiss R., 3rd Analysis of recombinant DNA using Escherichia coli minicells. Methods Enzymol. 1983;101:347–362. doi: 10.1016/0076-6879(83)01026-5. [DOI] [PubMed] [Google Scholar]
- Datta P., Goss T. J., Omnaas J. R., Patil R. V. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: homology with other dehydratases. Proc Natl Acad Sci U S A. 1987 Jan;84(2):393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaney A. D. A DNA sequence handling program. Nucleic Acids Res. 1982 Jan 11;10(1):61–67. doi: 10.1093/nar/10.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss T. J., Datta P. Escherichia coli K-12 mutation that inactivates biodegradative threonine dehydratase by transposon Tn5 insertion. J Bacteriol. 1984 Jun;158(3):826–831. doi: 10.1128/jb.158.3.826-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss T. J., Datta P. Molecular cloning and expression of the biodegradative threonine dehydratase gene (tdc) of Escherichia coli K12. Mol Gen Genet. 1985;201(2):308–314. doi: 10.1007/BF00425676. [DOI] [PubMed] [Google Scholar]
- Grantham R., Gautier C., Gouy M., Jacobzone M., Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981 Jan 10;9(1):r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert E. H., Datta P. Synthesis of biodegradative threonine dehydratase in Escherichia coli: role of amino acids, electron acceptors, and certain intermediary metabolites. J Bacteriol. 1983 Aug;155(2):586–592. doi: 10.1128/jb.155.2.586-592.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson T. J., Schumacher G., Boos W. Identification of the glpT-encoded sn-glycerol-3-phosphate permease of Escherichia coli, an oligomeric integral membrane protein. J Bacteriol. 1982 Dec;152(3):1008–1021. doi: 10.1128/jb.152.3.1008-1021.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merberg D., Datta P. Altered expression of biodegradative threonine dehydratase in Escherichia coli mutants. J Bacteriol. 1982 Apr;150(1):52–59. doi: 10.1128/jb.150.1.52-59.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Schweizer H., Boos W. Characterization of the ugp region containing the genes for the phoB dependent sn-glycerol-3-phosphate transport system of Escherichia coli. Mol Gen Genet. 1984;197(1):161–168. doi: 10.1007/BF00327937. [DOI] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbarger H. E. Amino acid biosynthesis and its regulation. Annu Rev Biochem. 1978;47:532–606. doi: 10.1146/annurev.bi.47.070178.002533. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yanofsky C., Platt T., Crawford I. P., Nichols B. P., Christie G. E., Horowitz H., VanCleemput M., Wu A. M. The complete nucleotide sequence of the tryptophan operon of Escherichia coli. Nucleic Acids Res. 1981 Dec 21;9(24):6647–6668. doi: 10.1093/nar/9.24.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]